精彩专栏推荐订阅:在下方主页👇🏻👇🏻👇🏻👇🏻
💖🔥作者主页 :计算机毕设木哥🔥 💖
文章目录
- 一、项目介绍
- 二、视频展示
- 三、开发环境
- 四、系统展示
- 五、代码展示
- 六、项目文档展示
- 七、项目总结
- [<font color=#fe2c24 >大家可以帮忙点赞、收藏、关注、评论啦 👇🏻](#fe2c24 >大家可以帮忙点赞、收藏、关注、评论啦 👇🏻)
一、项目介绍
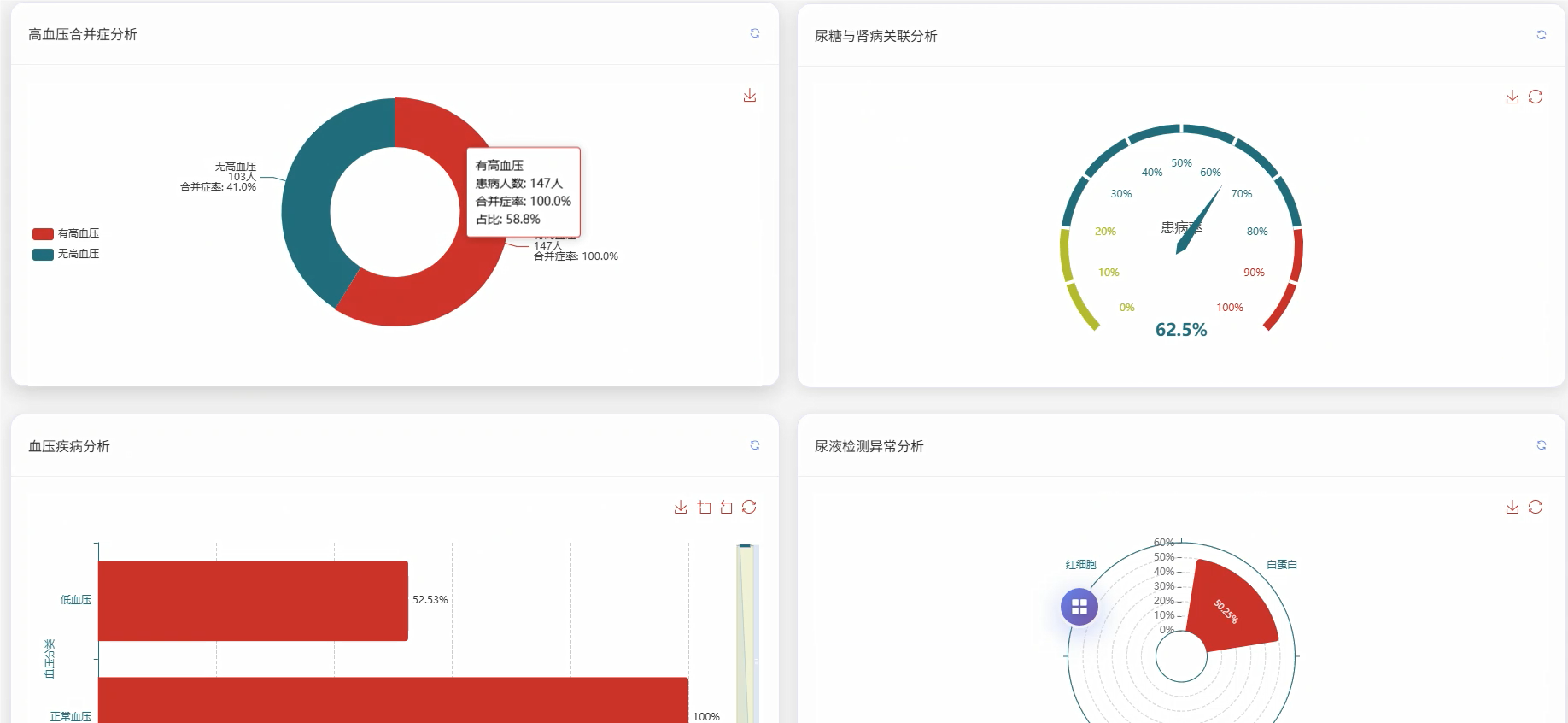
基于大数据的慢性肾病数据可视化分析系统是一套运用现代大数据技术栈构建的医疗数据分析平台,该系统采用Hadoop分布式存储架构结合Spark大数据处理引擎,能够高效处理海量慢性肾病患者的医疗数据。系统后端基于Python语言和Django框架搭建,前端采用Vue.js框架配合ECharts可视化库,为用户提供直观的数据展示界面。系统核心功能涵盖慢性肾病患病情况统计分析、肾功能指标深度分析、血液生化指标综合评估、多指标联合诊断价值分析、疾病进展与严重程度评估以及临床特征模式识别分析等六大模块。通过对血压、血尿素、血清肌酐、尿液检查等多维度医疗指标的智能分析,系统能够为医疗工作者提供患病率分布统计、高血压合并慢性肾病关联性分析、肾功能损害程度评估、电解质平衡监测等多项专业分析服务,同时支持疾病严重程度分层统计和并发症发生情况追踪,为临床诊断决策和疾病管理提供数据支撑。
选题背景
慢性肾病作为全球性的公共卫生问题,其患病率在世界范围内呈现持续上升的趋势,我国成年人群中慢性肾病的患病率已达到相当高的水平,给国家医疗体系带来了沉重负担。传统的慢性肾病诊断和监测主要依靠医生的临床经验和单一指标判断,这种方式在面对大量患者数据时存在效率低下、主观性强、易遗漏关键信息等问题。随着医疗信息化建设的推进,医院积累了大量的患者检查数据,这些数据蕴含着丰富的疾病发展规律和诊断信息,但由于缺乏有效的数据分析工具,这些宝贵的医疗数据往往未能得到充分利用。传统的数据库系统在处理海量医疗数据时面临性能瓶颈,难以满足实时分析的需求,而大数据技术的发展为解决这一问题提供了新的思路和技术支撑。
选题意义
本课题研究具有重要的实际应用价值和理论探索意义。从实际应用角度来看,该系统能够帮助医疗工作者更高效地处理和分析慢性肾病相关数据,通过多维度指标的综合分析提高疾病诊断的准确性和及时性,为临床决策提供科学依据。系统的可视化功能使得复杂的医疗数据能够以直观的图表形式呈现,降低了医务人员理解和使用数据的门槛,有助于提升医疗服务质量。对于患者而言,系统能够实现疾病风险的早期识别和预警,有助于疾病的早期干预和治疗效果的改善。从技术层面来说,本课题探索了大数据技术在医疗健康领域的具体应用模式,验证了Hadoop和Spark技术栈在处理医疗大数据场景下的可行性和有效性。通过将传统的医疗数据分析与现代大数据技术相结合,为相关领域的技术应用提供了一定的参考价值,同时也为计算机专业学生提供了一个贴近实际应用需求的实践平台。
二、视频展示
2026年计算机毕设推荐:基于大数据的慢性肾病数据可视化分析系统技术选型指南【Hadoop、spark、python】
三、开发环境
- 大数据技术:Hadoop、Spark、Hive
- 开发技术:Python、Django框架、Vue、Echarts
- 软件工具:Pycharm、DataGrip、Anaconda
- 可视化 工具 Echarts
四、系统展示
登录模块:
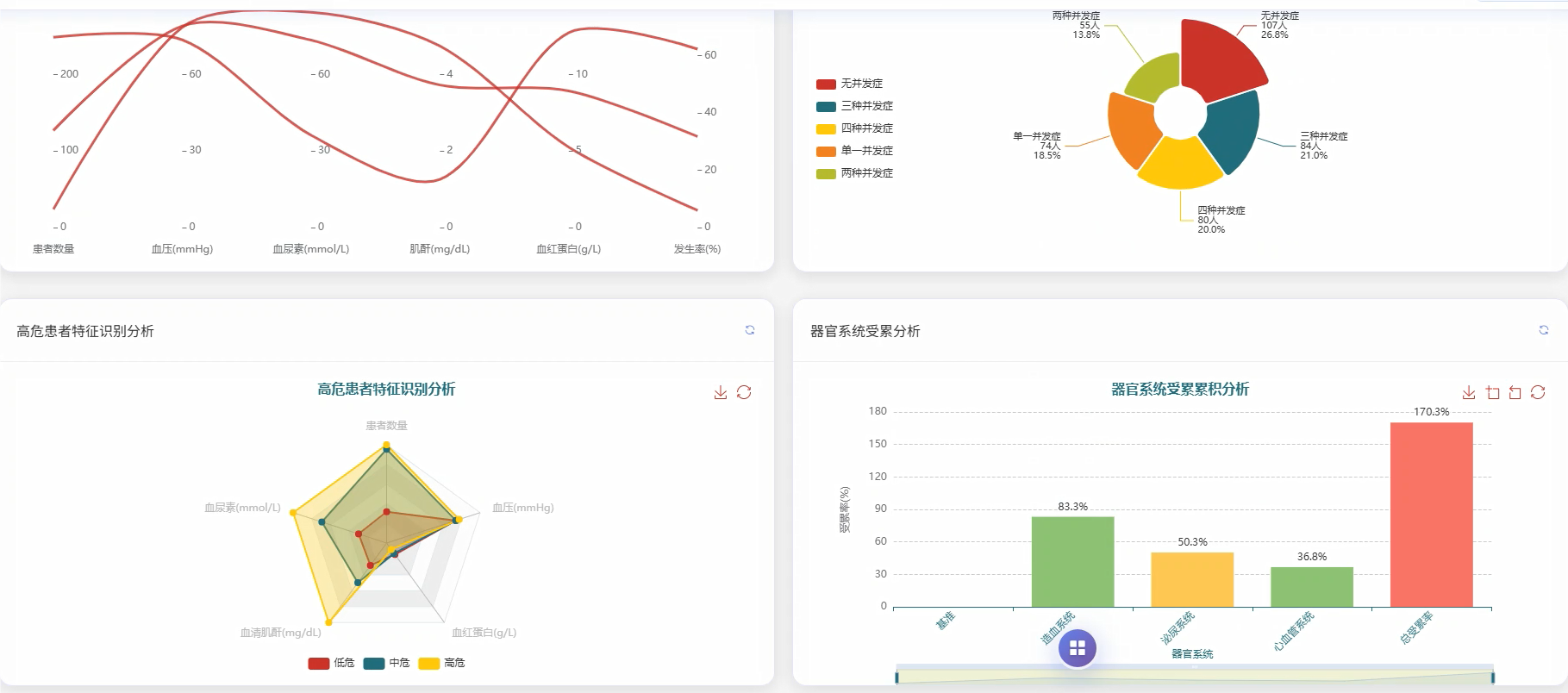
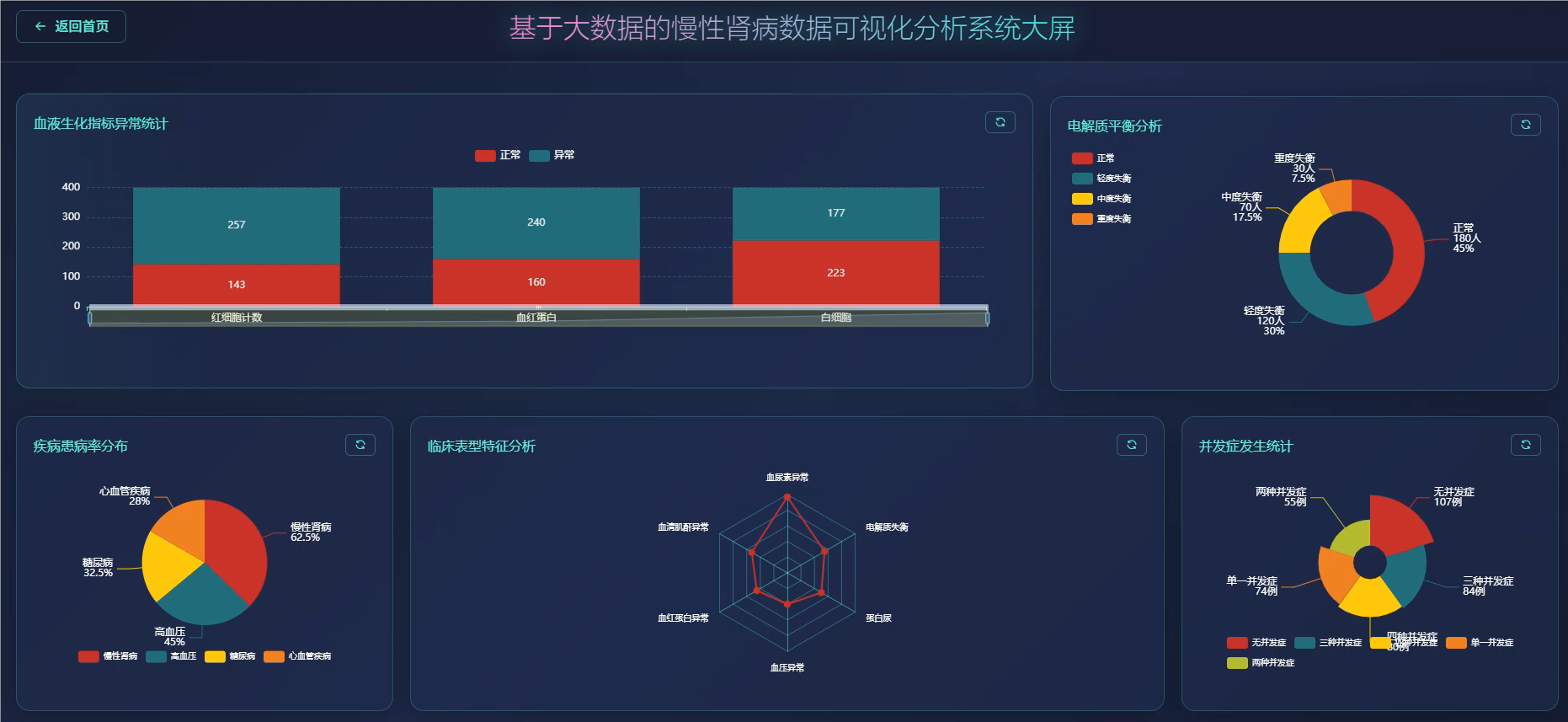
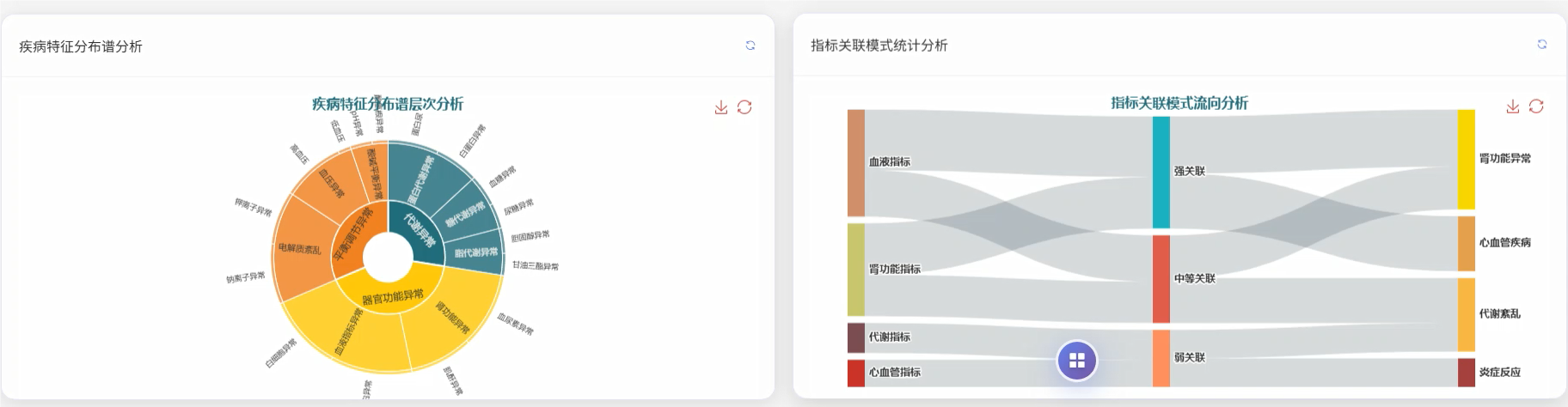
管理模块展示:
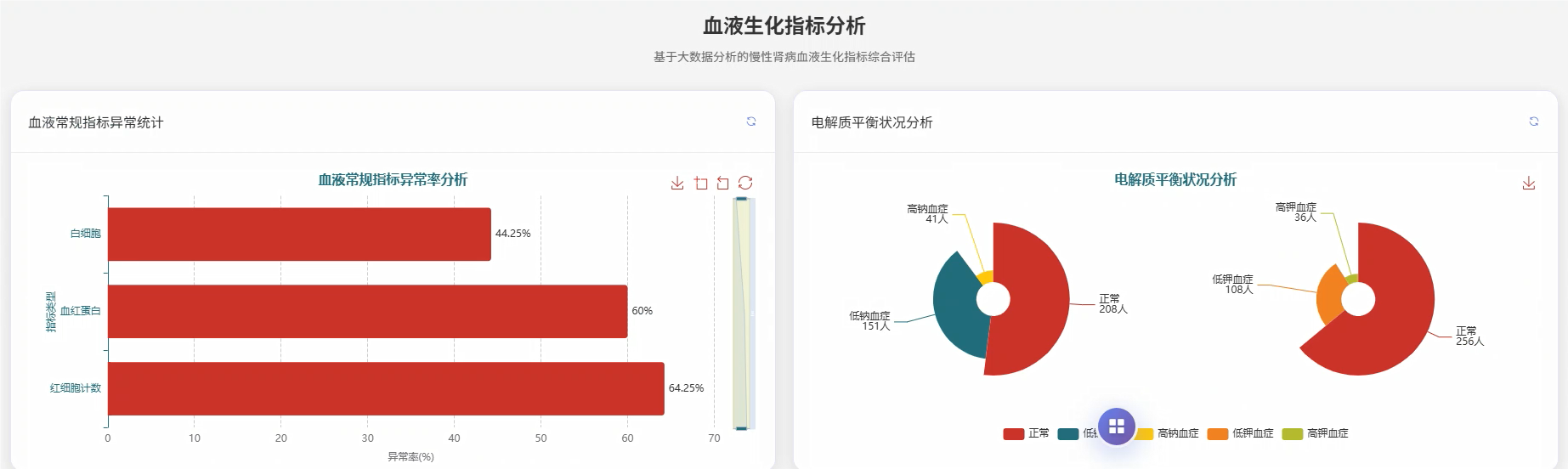
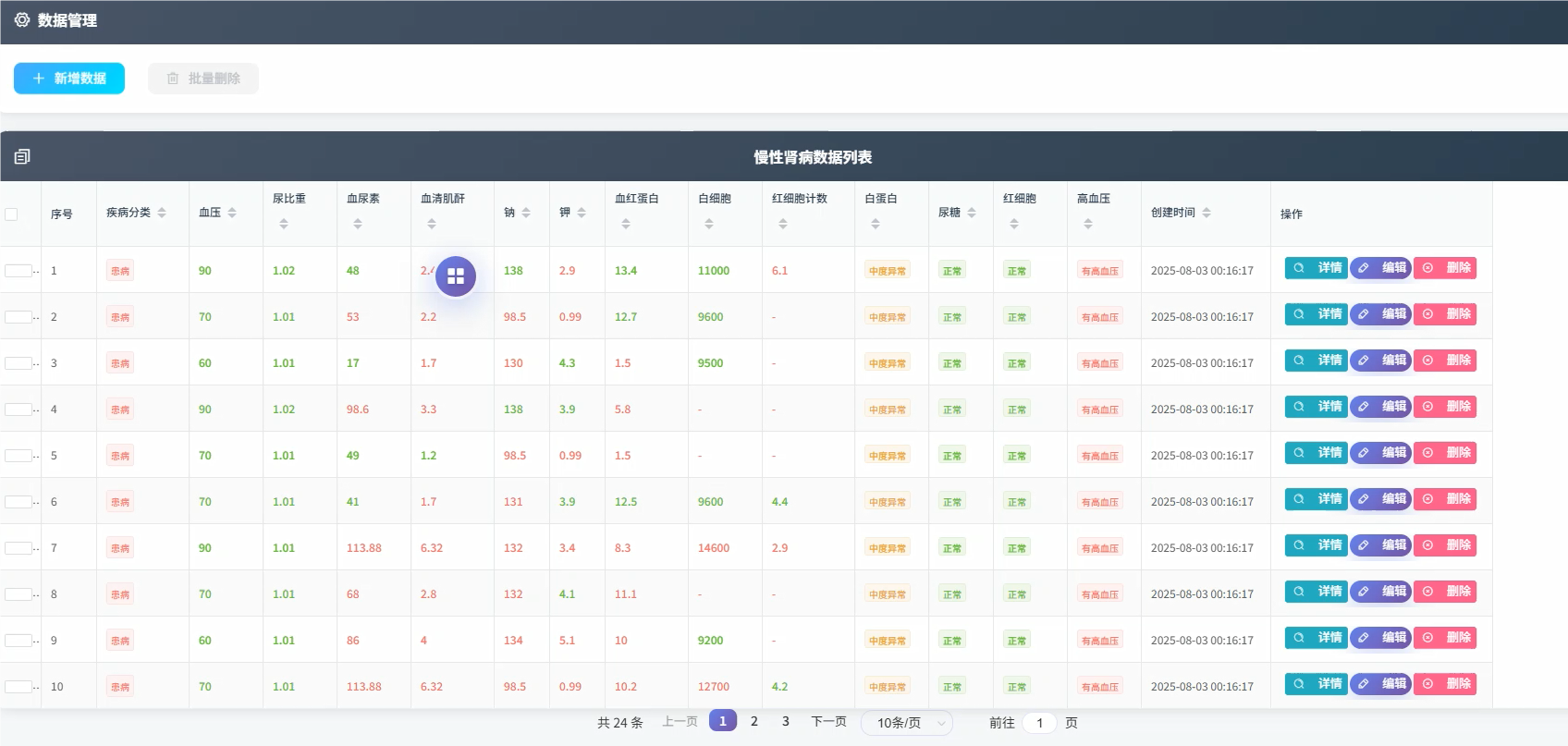
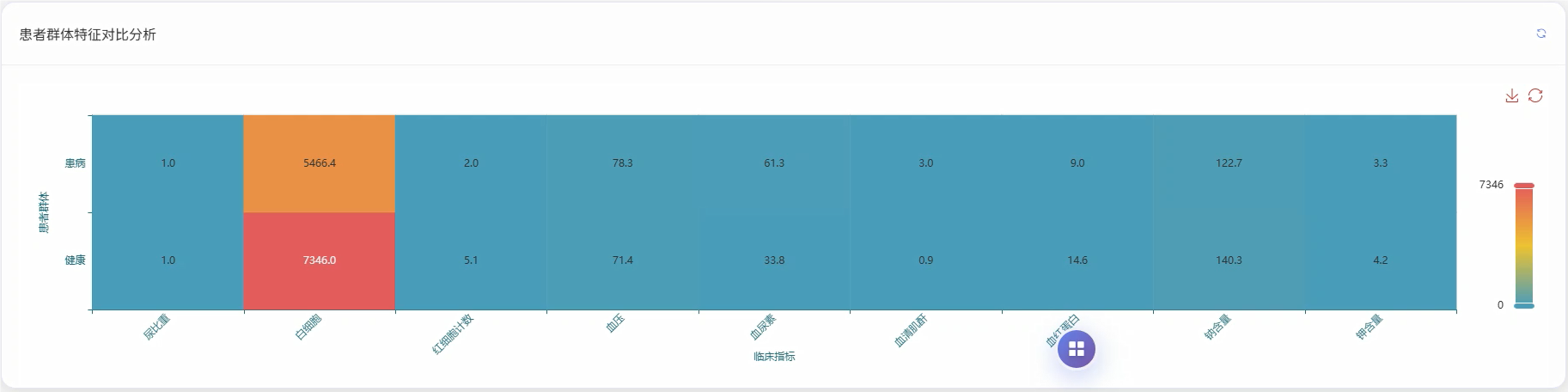
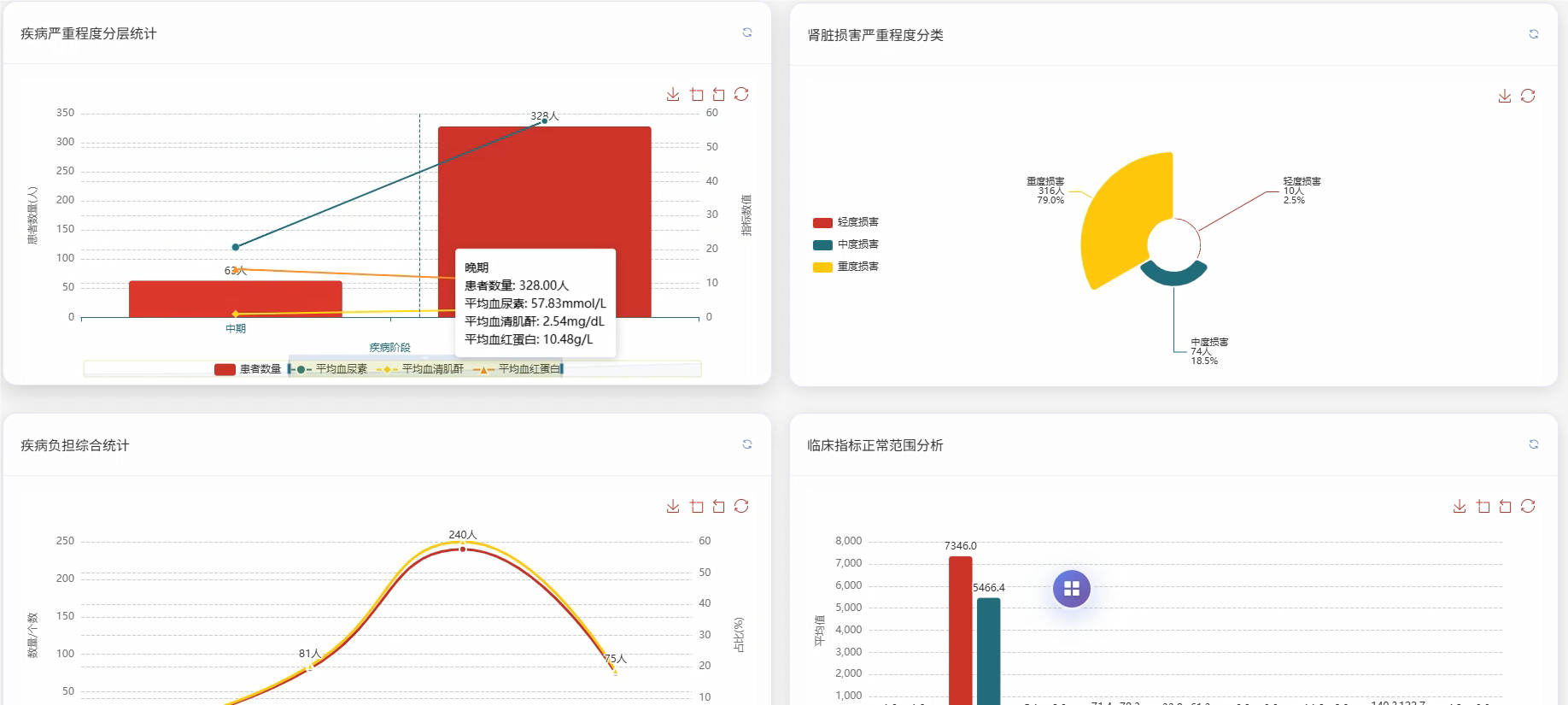
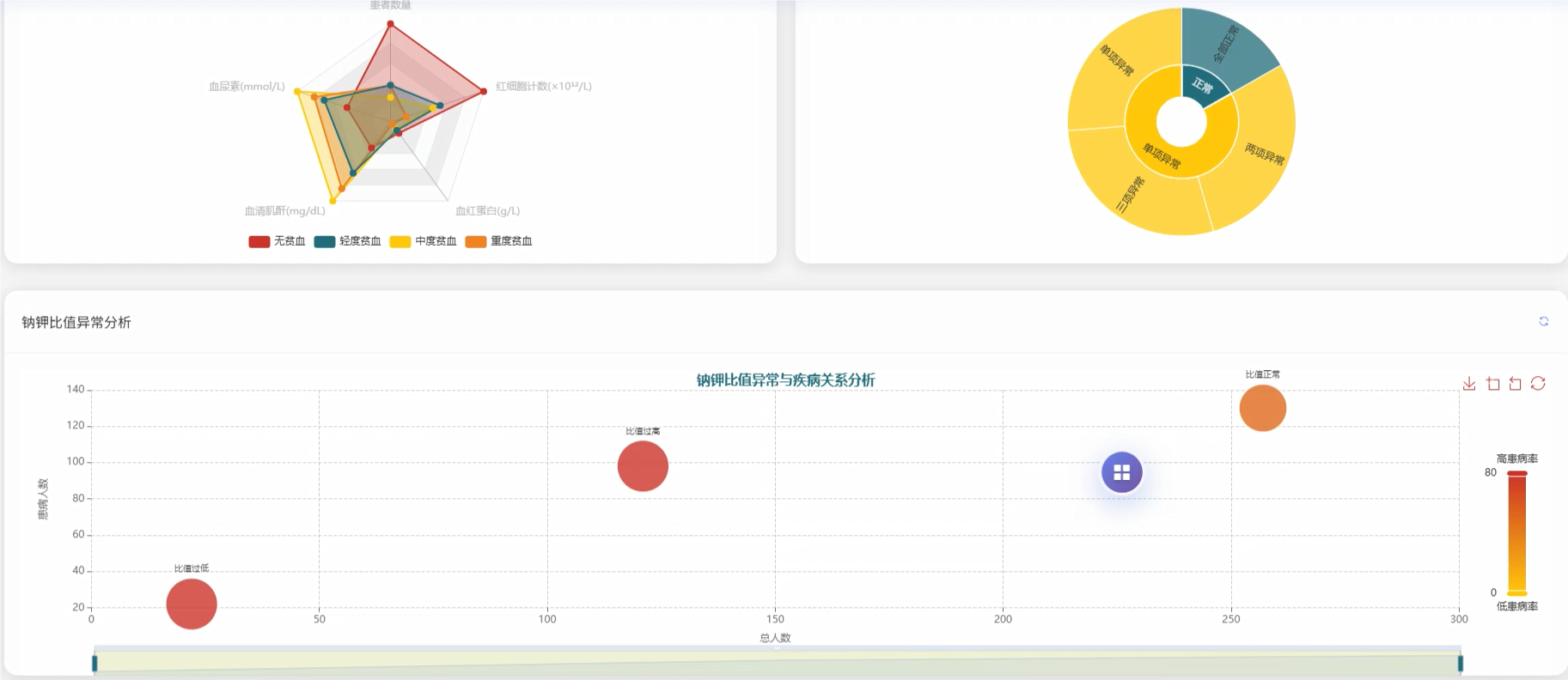
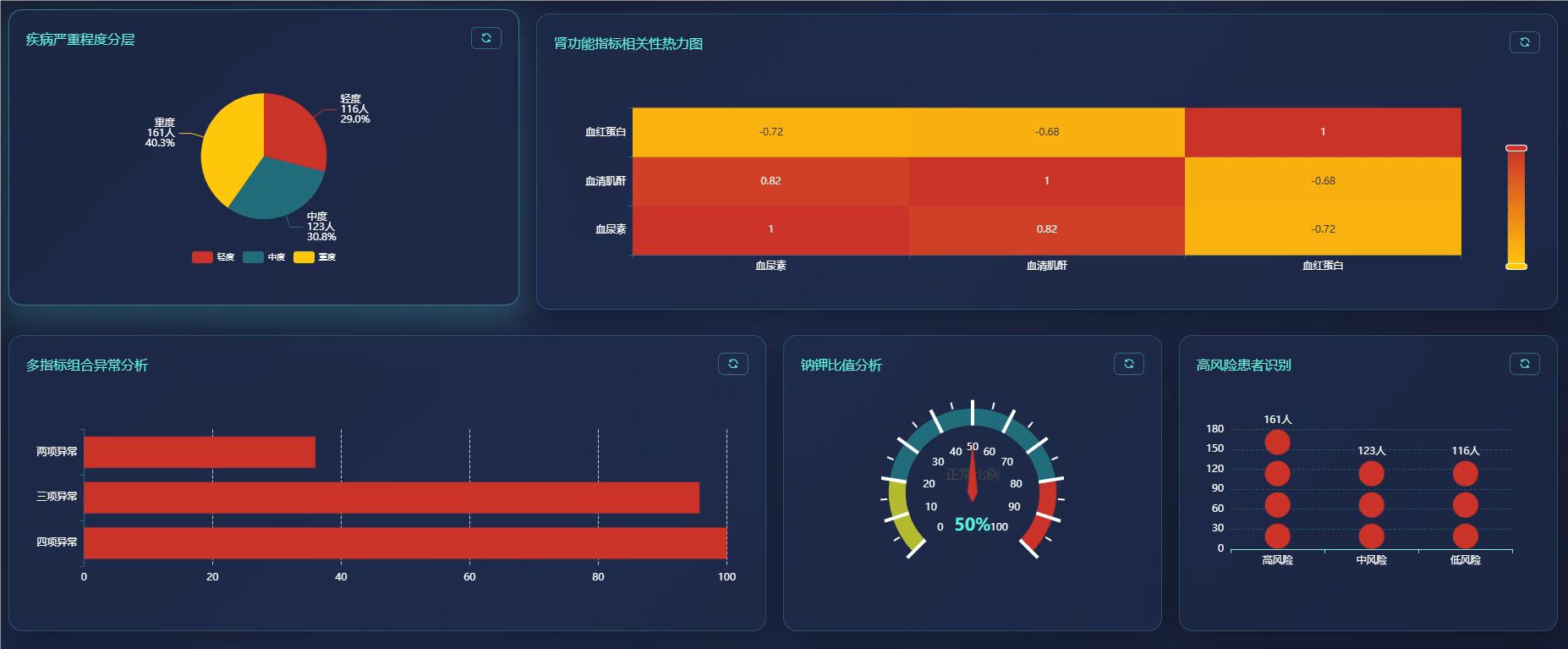
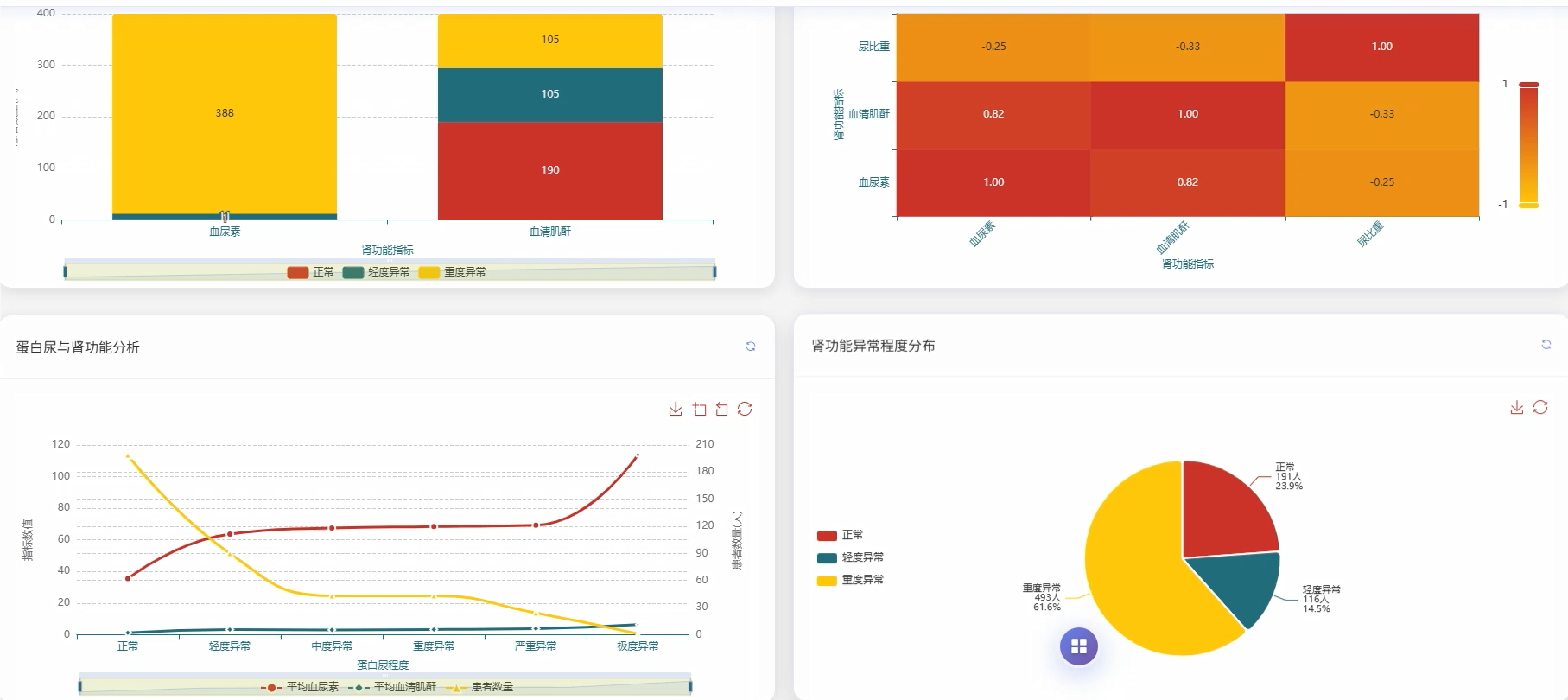
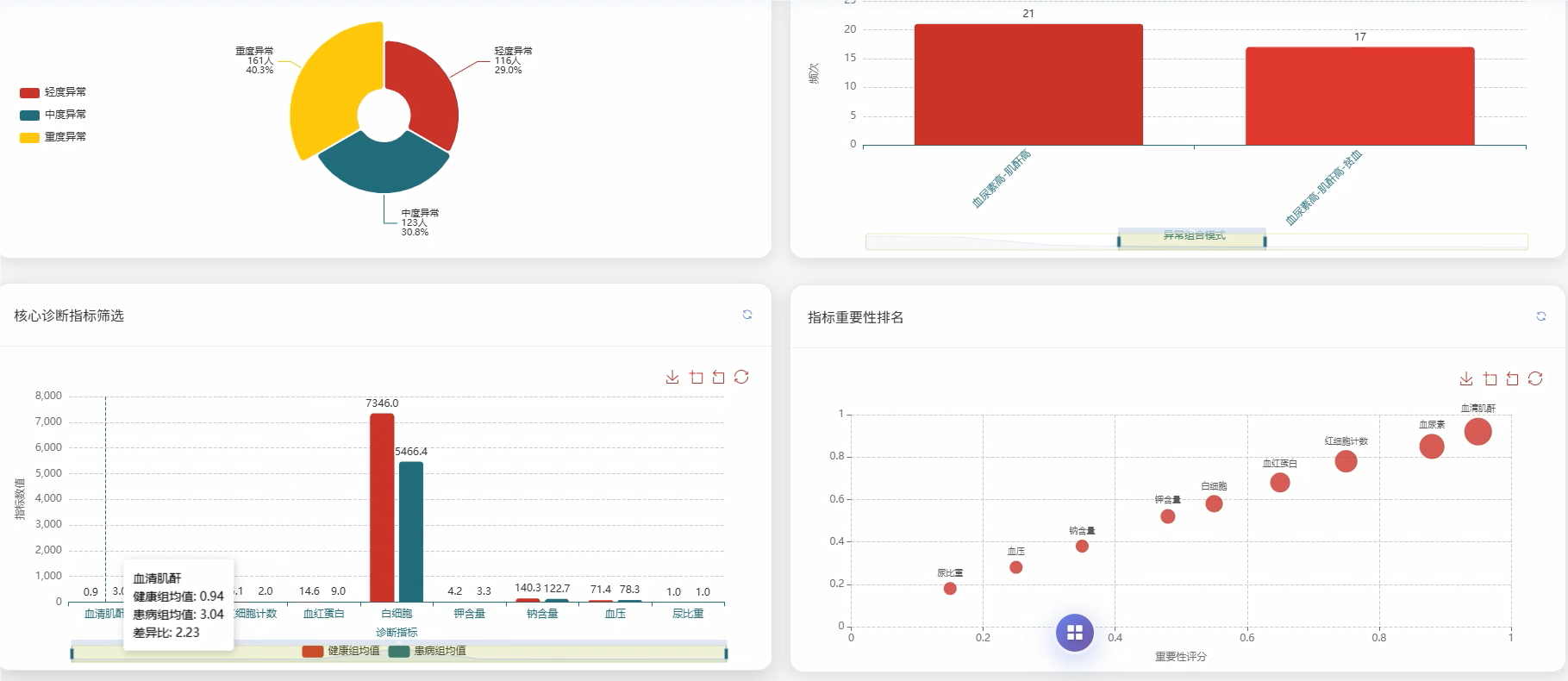
五、代码展示
bash
from pyspark.sql import SparkSession
from pyspark.sql.functions import col, when, count, avg, desc, asc
from django.http import JsonResponse
from django.views.decorators.csrf import csrf_exempt
import json
spark = SparkSession.builder.appName("ChronicKidneyDiseaseAnalysis").config("spark.executor.memory", "2g").getOrCreate()
@csrf_exempt
def analyze_disease_distribution(request):
if request.method == 'POST':
data = json.loads(request.body)
df = spark.read.option("header", "true").csv("/data/chronic_kidney_disease.csv")
df = df.withColumn("Bu", col("Bu").cast("double")).withColumn("Sc", col("Sc").cast("double")).withColumn("Bp", col("Bp").cast("double"))
total_count = df.count()
disease_stats = df.groupBy("Class").agg(count("*").alias("count")).collect()
healthy_count = next((row.count for row in disease_stats if row.Class == "notckd"), 0)
disease_count = next((row.count for row in disease_stats if row.Class == "ckd"), 0)
healthy_rate = round(healthy_count / total_count * 100, 2) if total_count > 0 else 0
disease_rate = round(disease_count / total_count * 100, 2) if total_count > 0 else 0
bp_analysis = df.withColumn("bp_level", when(col("Bp") < 120, "normal").when(col("Bp") < 140, "prehypertension").when(col("Bp") < 160, "stage1_hypertension").otherwise("stage2_hypertension"))
bp_disease_stats = bp_analysis.groupBy("bp_level", "Class").agg(count("*").alias("count")).collect()
bp_disease_dict = {}
for row in bp_disease_stats:
if row.bp_level not in bp_disease_dict:
bp_disease_dict[row.bp_level] = {"total": 0, "disease": 0}
bp_disease_dict[row.bp_level]["total"] += row.count
if row.Class == "ckd":
bp_disease_dict[row.bp_level]["disease"] += row.count
for level in bp_disease_dict:
total = bp_disease_dict[level]["total"]
disease = bp_disease_dict[level]["disease"]
bp_disease_dict[level]["disease_rate"] = round(disease / total * 100, 2) if total > 0 else 0
htn_analysis = df.groupBy("Htn", "Class").agg(count("*").alias("count")).collect()
htn_disease_dict = {}
for row in htn_analysis:
if row.Htn not in htn_disease_dict:
htn_disease_dict[row.Htn] = {"total": 0, "disease": 0}
htn_disease_dict[row.Htn]["total"] += row.count
if row.Class == "ckd":
htn_disease_dict[row.Htn]["disease"] += row.count
for htn_status in htn_disease_dict:
total = htn_disease_dict[htn_status]["total"]
disease = htn_disease_dict[htn_status]["disease"]
htn_disease_dict[htn_status]["disease_rate"] = round(disease / total * 100, 2) if total > 0 else 0
result_data = {"total_patients": total_count, "healthy_count": healthy_count, "disease_count": disease_count, "healthy_rate": healthy_rate, "disease_rate": disease_rate, "bp_analysis": bp_disease_dict, "htn_analysis": htn_disease_dict}
return JsonResponse(result_data)
@csrf_exempt
def analyze_kidney_function(request):
if request.method == 'POST':
data = json.loads(request.body)
df = spark.read.option("header", "true").csv("/data/chronic_kidney_disease.csv")
df = df.withColumn("Bu", col("Bu").cast("double")).withColumn("Sc", col("Sc").cast("double")).withColumn("Al", col("Al").cast("double")).withColumn("Sg", col("Sg").cast("double"))
bu_analysis = df.withColumn("bu_level", when(col("Bu") <= 20, "normal").when(col("Bu") <= 50, "mild_abnormal").otherwise("severe_abnormal"))
bu_stats = bu_analysis.groupBy("bu_level", "Class").agg(count("*").alias("count")).collect()
bu_distribution = {}
for row in bu_stats:
if row.bu_level not in bu_distribution:
bu_distribution[row.bu_level] = {"total": 0, "disease": 0, "healthy": 0}
bu_distribution[row.bu_level]["total"] += row.count
if row.Class == "ckd":
bu_distribution[row.bu_level]["disease"] += row.count
else:
bu_distribution[row.bu_level]["healthy"] += row.count
sc_analysis = df.withColumn("sc_level", when(col("Sc") <= 1.2, "normal").when(col("Sc") <= 3.0, "mild_abnormal").otherwise("severe_abnormal"))
sc_stats = sc_analysis.groupBy("sc_level", "Class").agg(count("*").alias("count")).collect()
sc_distribution = {}
for row in sc_stats:
if row.sc_level not in sc_distribution:
sc_distribution[row.sc_level] = {"total": 0, "disease": 0, "healthy": 0}
sc_distribution[row.sc_level]["total"] += row.count
if row.Class == "ckd":
sc_distribution[row.sc_level]["disease"] += row.count
else:
sc_distribution[row.sc_level]["healthy"] += row.count
al_analysis = df.withColumn("al_level", when(col("Al").isNull() | (col("Al") == 0), "normal").when(col("Al") <= 2, "mild_proteinuria").when(col("Al") <= 4, "moderate_proteinuria").otherwise("severe_proteinuria"))
al_stats = al_analysis.groupBy("al_level", "Class").agg(count("*").alias("count")).collect()
al_distribution = {}
for row in al_stats:
if row.al_level not in al_distribution:
al_distribution[row.al_level] = {"total": 0, "disease": 0, "healthy": 0}
al_distribution[row.al_level]["total"] += row.count
if row.Class == "ckd":
al_distribution[row.al_level]["disease"] += row.count
else:
al_distribution[row.al_level]["healthy"] += row.count
severity_analysis = df.withColumn("severity", when((col("Bu") <= 20) & (col("Sc") <= 1.2), "mild").when((col("Bu") <= 50) & (col("Sc") <= 3.0), "moderate").otherwise("severe"))
severity_stats = severity_analysis.groupBy("severity", "Class").agg(count("*").alias("count")).collect()
severity_distribution = {}
for row in severity_stats:
if row.severity not in severity_distribution:
severity_distribution[row.severity] = {"total": 0, "disease": 0}
severity_distribution[row.severity]["total"] += row.count
if row.Class == "ckd":
severity_distribution[row.severity]["disease"] += row.count
for level in severity_distribution:
total = severity_distribution[level]["total"]
disease = severity_distribution[level]["disease"]
severity_distribution[level]["disease_rate"] = round(disease / total * 100, 2) if total > 0 else 0
result_data = {"bu_distribution": bu_distribution, "sc_distribution": sc_distribution, "al_distribution": al_distribution, "severity_distribution": severity_distribution}
return JsonResponse(result_data)
@csrf_exempt
def analyze_blood_biochemistry(request):
if request.method == 'POST':
data = json.loads(request.body)
df = spark.read.option("header", "true").csv("/data/chronic_kidney_disease.csv")
df = df.withColumn("Hemo", col("Hemo").cast("double")).withColumn("Wbcc", col("Wbcc").cast("double")).withColumn("Rbcc", col("Rbcc").cast("double")).withColumn("Sod", col("Sod").cast("double")).withColumn("Pot", col("Pot").cast("double"))
hemo_analysis = df.withColumn("hemo_level", when(col("Hemo") >= 12.0, "normal").when(col("Hemo") >= 9.0, "mild_anemia").otherwise("severe_anemia"))
hemo_stats = hemo_analysis.groupBy("hemo_level", "Class").agg(count("*").alias("count")).collect()
hemo_distribution = {}
for row in hemo_stats:
if row.hemo_level not in hemo_distribution:
hemo_distribution[row.hemo_level] = {"total": 0, "disease": 0, "healthy": 0}
hemo_distribution[row.hemo_level]["total"] += row.count
if row.Class == "ckd":
hemo_distribution[row.hemo_level]["disease"] += row.count
else:
hemo_distribution[row.hemo_level]["healthy"] += row.count
wbc_analysis = df.withColumn("wbc_level", when((col("Wbcc") >= 4000) & (col("Wbcc") <= 11000), "normal").when(col("Wbcc") < 4000, "leukopenia").otherwise("leukocytosis"))
wbc_stats = wbc_analysis.groupBy("wbc_level", "Class").agg(count("*").alias("count")).collect()
wbc_distribution = {}
for row in wbc_stats:
if row.wbc_level not in wbc_distribution:
wbc_distribution[row.wbc_level] = {"total": 0, "disease": 0, "healthy": 0}
wbc_distribution[row.wbc_level]["total"] += row.count
if row.Class == "ckd":
wbc_distribution[row.wbc_level]["disease"] += row.count
else:
wbc_distribution[row.wbc_level]["healthy"] += row.count
electrolyte_analysis = df.withColumn("sod_level", when((col("Sod") >= 135) & (col("Sod") <= 145), "normal").when(col("Sod") < 135, "hyponatremia").otherwise("hypernatremia"))
electrolyte_analysis = electrolyte_analysis.withColumn("pot_level", when((col("Pot") >= 3.5) & (col("Pot") <= 5.0), "normal").when(col("Pot") < 3.5, "hypokalemia").otherwise("hyperkalemia"))
sod_stats = electrolyte_analysis.groupBy("sod_level", "Class").agg(count("*").alias("count")).collect()
sod_distribution = {}
for row in sod_stats:
if row.sod_level not in sod_distribution:
sod_distribution[row.sod_level] = {"total": 0, "disease": 0, "healthy": 0}
sod_distribution[row.sod_level]["total"] += row.count
if row.Class == "ckd":
sod_distribution[row.sod_level]["disease"] += row.count
else:
sod_distribution[row.sod_level]["healthy"] += row.count
pot_stats = electrolyte_analysis.groupBy("pot_level", "Class").agg(count("*").alias("count")).collect()
pot_distribution = {}
for row in pot_stats:
if row.pot_level not in pot_distribution:
pot_distribution[row.pot_level] = {"total": 0, "disease": 0, "healthy": 0}
pot_distribution[row.pot_level]["total"] += row.count
if row.Class == "ckd":
pot_distribution[row.pot_level]["disease"] += row.count
else:
pot_distribution[row.pot_level]["healthy"] += row.count
anemia_kidney_correlation = df.filter((col("Hemo").isNotNull()) & (col("Bu").isNotNull()) & (col("Sc").isNotNull()))
anemia_kidney_stats = anemia_kidney_correlation.withColumn("anemia_kidney_risk", when((col("Hemo") < 12.0) & (col("Bu") > 20), "high_risk").when((col("Hemo") < 12.0) | (col("Bu") > 20), "medium_risk").otherwise("low_risk"))
risk_stats = anemia_kidney_stats.groupBy("anemia_kidney_risk", "Class").agg(count("*").alias("count")).collect()
risk_distribution = {}
for row in risk_stats:
if row.anemia_kidney_risk not in risk_distribution:
risk_distribution[row.anemia_kidney_risk] = {"total": 0, "disease": 0}
risk_distribution[row.anemia_kidney_risk]["total"] += row.count
if row.Class == "ckd":
risk_distribution[row.anemia_kidney_risk]["disease"] += row.count
for risk_level in risk_distribution:
total = risk_distribution[risk_level]["total"]
disease = risk_distribution[risk_level]["disease"]
risk_distribution[risk_level]["disease_rate"] = round(disease / total * 100, 2) if total > 0 else 0
result_data = {"hemo_distribution": hemo_distribution, "wbc_distribution": wbc_distribution, "sod_distribution": sod_distribution, "pot_distribution": pot_distribution, "anemia_kidney_risk": risk_distribution}
return JsonResponse(result_data)六、项目文档展示

七、项目总结
本课题成功构建了基于大数据技术的慢性肾病数据可视化分析系统,该系统充分运用了Hadoop分布式存储和Spark大数据处理技术的优势,为海量医疗数据的高效分析提供了可行的技术方案。通过采用Python语言和Django框架搭建后端服务,结合Vue.js和ECharts技术实现前端数据可视化,系统在技术架构上体现了现代大数据应用的典型特征。在功能实现方面,系统涵盖了慢性肾病诊断所需的核心数据分析模块,包括患病情况统计分析、肾功能指标评估、血液生化指标监测等多个维度的综合分析功能,能够为医疗工作者提供科学的数据支撑。系统的可视化界面使得复杂的医疗数据能够以直观的图表形式呈现,有效降低了数据理解和使用的门槛。通过本课题的实践,验证了大数据技术在医疗健康领域应用的可行性,为类似的医疗数据分析系统开发提供了一定的技术参考。虽然作为毕业设计项目,系统在功能深度和数据处理规模上还有进一步优化的空间,但在技术选型和系统架构设计上体现了良好的工程实践价值。
大家可以帮忙点赞、收藏、关注、评论啦 👇🏻
💖🔥作者主页 :计算机毕设木哥🔥 💖