
前言
模型的损失计算包括3个方面,分别是:
- 定位损失
- 分类损失
- 置信度损失
损失的计算公式如下:
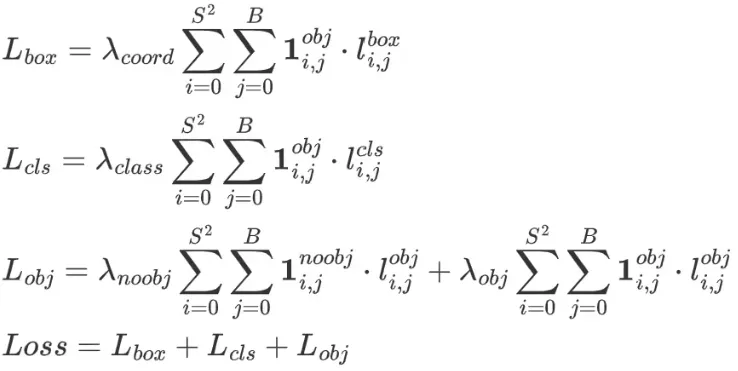
损失计算的代码流程也是按照这三大块来计算的。本篇主要讲解yolov5中损失计算的实现,包括损失的逻辑实现,张量操作的细节等。
准备工作
初始化损失张量的值,获取正样本的信息。
python
lcls = torch.zeros(1, device=self.device) # class loss
lbox = torch.zeros(1, device=self.device) # box loss
lobj = torch.zeros(1, device=self.device) # object loss
tcls, tbox, indices, anchors = self.build_targets(p, targets, imgs) # targets其中获取正样本信息在前面一篇文章中已经详细的分析过流程。这里返回的结果分别是:
- tcls:保存类别id
- tbox:保存的是gt中心相对于所在grid cell左上角偏移量
- indices:image_id, anchor_id, grid x刻度 grid y刻度
- anchors:保存anchor的具体宽高
python
tcls, tbox, indices, anchors = self.build_targets(p, targets, imgs) # targets遍历3种尺度,获取对应正样本的image_id,anchor_id,网格坐标xy。计算损失的大体思路是将3种尺度的损失值加在一起,所以这里分别处理每一种尺度。
python
for i, pi in enumerate(p): # layer index, layer predictions
b, a, gj, gi = indices[i] # image, anchor, gridy, gridx
tobj = torch.zeros(pi.shape[:4], dtype=pi.dtype, device=self.device) # target obj
n = b.shape[0] # number of targets从模型输出中筛选出正样本对应网格
python
b, a, gj, gi = indices[i]
pxy, pwh, _, pcls = pi[b, a, gj, gi].split((2, 2, 1, self.nc), 1) # target-subset of predictionspi 的形状是 6 * 3 * 80 * 80 * 7,pi是模型推理的输出结果,代表着 6张图片,一张图片中有3种anchor的结果,每一个anchor下是 80 * 80 的网格,每一个网格下的结果有7个输出,分别是nc=5,类别数2。5是xywh+confidence。
python
(Pdb) pp pi.shape
torch.Size([6, 3, 80, 80, 7])另外b、a、gj、gi等都是索引下标
b: image_id
a: anchor_id
gj: 网格y轴
gi: 网格x轴
python
Pdb) pp b
tensor([0, 0, 0, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 3, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 5, 0, 0, 0, 0, 0, 0, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 5, 5, 5, 0, 0, 0, 0, 0, 0, 0, 1, 1, 2, 2, 2, 2,
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 5, 5, 5, 5, 5, 0, 2, 2, 2, 2, 2, 2, 2, 3, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 5, 0, 1, 2, 2, 2, 2, 2, 2, 2, 2, 3, 3, 3, 3, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 5, 0, 0, 1, 2, 2,
2, 2, 2, 2, 2, 2, 3, 3, 3, 3, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 5, 0, 2, 2, 2, 2, 2, 2, 2, 2, 3, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 5, 0, 0, 0, 1, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 3, 3, 3, 3, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 5, 5, 0, 0, 1, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 3, 3, 3, 3, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4,
4, 4, 5, 5, 5, 0, 2, 2, 2, 2, 2, 4, 4, 4, 4, 4, 4, 4, 0, 0, 0, 0, 1, 2, 2, 2, 2, 2, 2, 3, 3, 3, 3, 3, 3, 3, 4, 4, 4, 4, 4, 4, 4, 4, 5, 5, 0, 0, 0, 0, 0, 1, 2, 2, 2, 2, 2, 2, 2, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 4, 4, 4, 4, 4, 4, 4, 4, 5, 5, 5, 5, 0, 0, 2, 2, 2, 2, 4, 4, 4, 4, 4, 4, 4, 0, 0, 0, 1, 2, 2, 2, 2, 3, 3,
3, 3, 3, 3, 3, 4, 4, 4, 4, 4, 4, 4, 4, 4, 5, 0, 0, 0, 0, 0, 1, 2, 2, 2, 2, 2, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 4, 4, 4, 4, 4, 4, 4, 4, 4, 5, 5], device='cuda:0')
(Pdb) pp a
tensor([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2,
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2,
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2,
2, 2, 2, 2, 2, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2], device='cuda:0')
(Pdb) pp gj
tensor([78, 74, 76, 13, 53, 54, 53, 53, 52, 52, 51, 50, 52, 52, 52, 76, 18, 18, 19, 17, 17, 18, 19, 17, 17, 17, 20, 19, 18, 18, 19, 23, 18, 41, 19, 39, 66, 67, 78, 74, 76, 15, 20, 13, 19, 57, 53, 54, 53, 53, 52, 52, 51, 50, 52, 52, 52, 31, 34, 34, 29, 34, 35, 29, 34, 76, 74, 74, 15, 14, 18, 18, 19, 17, 17, 18, 19, 17,
17, 17, 20, 19, 18, 18, 19, 23, 18, 41, 26, 32, 19, 42, 39, 42, 66, 67, 78, 74, 15, 20, 13, 19, 19, 57, 53, 54, 53, 53, 52, 52, 51, 50, 52, 52, 52, 31, 37, 34, 34, 29, 34, 35, 29, 34, 76, 74, 74, 75, 75, 15, 14, 18, 18, 19, 17, 17, 18, 19, 17, 17, 17, 20, 19, 18, 18, 19, 23, 18, 41, 46, 4, 31, 26, 32, 19, 74,
53, 53, 52, 51, 52, 52, 52, 76, 18, 19, 17, 17, 19, 17, 17, 20, 19, 19, 23, 19, 74, 15, 19, 53, 53, 52, 51, 52, 52, 52, 34, 29, 34, 76, 15, 18, 19, 17, 17, 19, 17, 17, 20, 19, 19, 23, 19, 42, 74, 15, 19, 53, 53, 52, 51, 52, 52, 52, 34, 29, 34, 76, 15, 18, 19, 17, 17, 19, 17, 17, 20, 19, 19, 23, 46, 19, 75, 52,
53, 52, 52, 51, 51, 49, 51, 75, 17, 17, 18, 16, 16, 18, 16, 19, 17, 18, 17, 18, 38, 65, 75, 14, 18, 56, 52, 53, 52, 52, 51, 51, 49, 51, 30, 28, 28, 75, 17, 17, 18, 16, 16, 18, 16, 19, 17, 18, 17, 31, 18, 38, 65, 14, 18, 56, 52, 53, 52, 52, 51, 51, 49, 51, 30, 28, 28, 75, 17, 17, 18, 16, 16, 18, 16, 19, 17, 18,
17, 45, 3, 31, 18, 78, 13, 54, 53, 52, 50, 18, 18, 17, 18, 18, 18, 41, 39, 66, 67, 78, 20, 13, 57, 54, 53, 52, 50, 31, 34, 34, 35, 29, 74, 74, 14, 18, 18, 17, 18, 18, 18, 41, 26, 32, 39, 42, 66, 67, 78, 20, 13, 19, 57, 54, 53, 52, 50, 31, 37, 34, 34, 35, 29, 74, 74, 75, 75, 14, 18, 18, 17, 18, 18, 18, 41, 4,
31, 26, 32, 79, 75, 14, 52, 53, 53, 19, 18, 18, 20, 19, 24, 42, 68, 79, 75, 21, 14, 52, 53, 53, 35, 35, 35, 36, 35, 75, 75, 16, 15, 19, 18, 18, 20, 19, 24, 42, 27, 43, 43, 68, 79, 75, 21, 14, 20, 52, 53, 53, 38, 35, 35, 35, 36, 35, 75, 75, 76, 76, 16, 15, 19, 18, 18, 20, 19, 24, 42, 32, 27], device='cuda:0')
(Pdb) pp gi
tensor([26, 73, 79, 51, 10, 16, 21, 25, 30, 36, 42, 44, 47, 52, 57, 56, 54, 24, 28, 33, 36, 40, 46, 60, 57, 52, 50, 48, 44, 37, 35, 39, 42, 6, 71, 50, 56, 11, 26, 73, 79, 10, 15, 51, 45, 62, 10, 16, 21, 25, 30, 36, 42, 44, 47, 52, 57, 62, 73, 79, 22, 1, 35, 13, 8, 56, 71, 2, 18, 3, 54, 24, 28, 33, 36, 40, 46, 60,
57, 52, 50, 48, 44, 37, 35, 39, 42, 6, 26, 10, 71, 9, 50, 39, 56, 11, 26, 73, 10, 15, 51, 45, 35, 62, 10, 16, 21, 25, 30, 36, 42, 44, 47, 52, 57, 62, 52, 73, 79, 22, 1, 35, 13, 8, 56, 71, 2, 20, 33, 18, 3, 54, 24, 28, 33, 36, 40, 46, 60, 57, 52, 50, 48, 44, 37, 35, 39, 42, 6, 41, 71, 45, 26, 10, 71, 72,
9, 20, 29, 41, 46, 51, 56, 55, 23, 27, 32, 35, 45, 59, 51, 49, 47, 34, 38, 70, 72, 9, 44, 9, 20, 29, 41, 46, 51, 56, 78, 21, 7, 55, 17, 23, 27, 32, 35, 45, 59, 51, 49, 47, 34, 38, 70, 8, 72, 9, 44, 9, 20, 29, 41, 46, 51, 56, 78, 21, 7, 55, 17, 23, 27, 32, 35, 45, 59, 51, 49, 47, 34, 38, 40, 70, 79, 10,
16, 21, 25, 30, 36, 44, 57, 56, 54, 24, 28, 33, 36, 46, 57, 50, 44, 35, 42, 71, 50, 56, 79, 10, 45, 62, 10, 16, 21, 25, 30, 36, 44, 57, 62, 22, 13, 56, 54, 24, 28, 33, 36, 46, 57, 50, 44, 35, 42, 10, 71, 50, 56, 10, 45, 62, 10, 16, 21, 25, 30, 36, 44, 57, 62, 22, 13, 56, 54, 24, 28, 33, 36, 46, 57, 50, 44, 35,
42, 41, 71, 10, 71, 27, 52, 17, 26, 37, 45, 55, 41, 58, 45, 38, 43, 7, 51, 57, 12, 27, 16, 52, 63, 17, 26, 37, 45, 63, 74, 2, 36, 14, 72, 3, 4, 55, 41, 58, 45, 38, 43, 7, 27, 11, 51, 40, 57, 12, 27, 16, 52, 36, 63, 17, 26, 37, 45, 63, 53, 74, 2, 36, 14, 72, 3, 21, 34, 4, 55, 41, 58, 45, 38, 43, 7, 72,
46, 27, 11, 26, 73, 51, 42, 47, 52, 40, 60, 52, 48, 37, 39, 6, 11, 26, 73, 15, 51, 42, 47, 52, 73, 79, 1, 35, 8, 71, 2, 18, 3, 40, 60, 52, 48, 37, 39, 6, 26, 9, 39, 11, 26, 73, 15, 51, 35, 42, 47, 52, 52, 73, 79, 1, 35, 8, 71, 2, 20, 33, 18, 3, 40, 60, 52, 48, 37, 39, 6, 45, 26], device='cuda:0')通过这四个张量的索引,就能得到所有模型输出结果中的具体定位信息xywh+confidence+两个类别。
split 将结果拆分,分别得到pxy:预测中心点 pwh:预测宽高 pcls:预测类别
python
pxy, pwh, _, pcls = pi[b, a, gj, gi].split((2, 2, 1, self.nc), 1) 因为b、a、gj、gi 是筛选出来的正样本,所以通过这一步就是获得了模型输出结果中,正样本网格对应的预测结果。方便后面和正样本中心点偏移量计算损失。
将原始预测信息还原成真实值
模型输出的结果是原始的预测结果,再计算损失值之前还需要将原始预测信息还原成真实值即boundingbox。
yolov5中计算预测真实值boundingbox的公式如下:
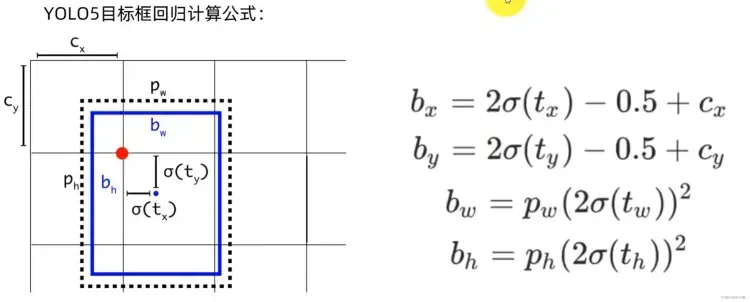
对应代码中首先将网络模型输出的结果通过sigmoid函数压缩到0-1之间,然后做对应的变换。
python
pxy = pxy.sigmoid() * 2 - 0.5
pwh = (pwh.sigmoid() * 2) ** 2 * anchors[i]
pbox = torch.cat((pxy, pwh), 1) # predicted boxpxy.sigmoid() * 2 - 0.5中pxy.sigmoid()取值范围为[0,1],那么pxy 变换结果的范围是[-0.5, 1.5],这表明坐标点可以超越该grid cell,出现在网格以外。这么设计的原因是当目标落在边界上时往往预测会比较困难,因为将参数回归到0或1比较难。但是如果预测的极限不在0,1,而是-0.5,1.5,那么边界上的预测就相对容易。
(pwh.sigmoid() * 2) ** 2 变换结果的范围是[0, 4],表示可以预测的范围是anchor的宽高的0倍至4倍。
注意:中心点xy的坐标是相对网格左上角的偏移量,而宽高的取值是相对anchor的比例。
定位损失
yolov5中定位损失使用的是CIOU 。关于CIOU损失的详解可以参考这篇文章
优化改进YOLOv5算法之添加GIoU、DIoU、CIoU、EIoU、Wise-IoU模块(超详细)_yolov5使用giou-CSDN博客
python
iou = bbox_iou(pbox, tbox[i], CIoU=True).squeeze() # iou(prediction, target)
lbox += (1.0 - iou).mean() # iou lossCIOU的计算公式如下:
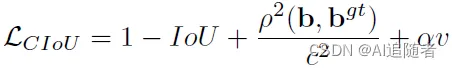
iou张量计算完成之后,再用1减去iou,去均值得到损失值。理解上面的公式再看下面的代码就会发现代码其实就是实现了公式的逻辑。
具体iou的计算过程如下:
python
def bbox_iou(box1, box2, x1y1x2y2=True, GIoU=False, DIoU=False, CIoU=False, eps=1e-7):
# Returns the IoU of box1 to box2. box1 is 4, box2 is nx4
box2 = box2.T
# Get the coordinates of bounding boxes
if x1y1x2y2: # x1, y1, x2, y2 = box1
b1_x1, b1_y1, b1_x2, b1_y2 = box1[0], box1[1], box1[2], box1[3]
b2_x1, b2_y1, b2_x2, b2_y2 = box2[0], box2[1], box2[2], box2[3]
else: # transform from xywh to xyxy
b1_x1, b1_x2 = box1[0] - box1[2] / 2, box1[0] + box1[2] / 2
b1_y1, b1_y2 = box1[1] - box1[3] / 2, box1[1] + box1[3] / 2
b2_x1, b2_x2 = box2[0] - box2[2] / 2, box2[0] + box2[2] / 2
b2_y1, b2_y2 = box2[1] - box2[3] / 2, box2[1] + box2[3] / 2
# Intersection area 交集
inter = (torch.min(b1_x2, b2_x2) - torch.max(b1_x1, b2_x1)).clamp(0) * \
(torch.min(b1_y2, b2_y2) - torch.max(b1_y1, b2_y1)).clamp(0)
# Union Area 并集
w1, h1 = b1_x2 - b1_x1, b1_y2 - b1_y1 + eps
w2, h2 = b2_x2 - b2_x1, b2_y2 - b2_y1 + eps
union = w1 * h1 + w2 * h2 - inter + eps
# 得到交并比IOU
iou = inter / union
if GIoU or DIoU or CIoU:
cw = torch.max(b1_x2, b2_x2) - torch.min(b1_x1, b2_x1) # convex (smallest enclosing box) width
ch = torch.max(b1_y2, b2_y2) - torch.min(b1_y1, b2_y1) # convex height
if CIoU or DIoU: # Distance or Complete IoU https://arxiv.org/abs/1911.08287v1
c2 = cw ** 2 + ch ** 2 + eps # convex diagonal squared
rho2 = ((b2_x1 + b2_x2 - b1_x1 - b1_x2) ** 2 +
(b2_y1 + b2_y2 - b1_y1 - b1_y2) ** 2) / 4 # center distance squared
if DIoU:
return iou - rho2 / c2 # DIoU
elif CIoU: # https://github.com/Zzh-tju/DIoU-SSD-pytorch/blob/master/utils/box/box_utils.py#L47
v = (4 / math.pi ** 2) * torch.pow(torch.atan(w2 / h2) - torch.atan(w1 / h1), 2)
with torch.no_grad():
alpha = v / (v - iou + (1 + eps))
return iou - (rho2 / c2 + v * alpha) # CIoU
else: # GIoU https://arxiv.org/pdf/1902.09630.pdf
c_area = cw * ch + eps # convex area
return iou - (c_area - union) / c_area # GIoU
else:
return iou # IoU置信度损失
置信度损失的一个特点就是需要正负样本都参与计算,所以正样本和负样本都有置信度。
- 正样本置信度:用预测框和标注框的DIOU结果作为正样本置信度
- 负样本置信度:为0
复制iou张量,生成一个不参与梯度计算的张量,并将iou的值都限制在0以上,小于0的会赋值为0,最后将所有元素的类型修改成和tobj一致。
python
iou = iou.detach().clamp(0).type(tobj.dtype)对iou排序,排序之后 如果同一个grid出现两个gt 那么我们经过排序之后每个grid中的score_iou都能保证是最大的。(小的会被覆盖 因为同一个grid坐标肯定相同)。那么从时间顺序的话, 最后1个总是和最大的IOU去计算LOSS
python
if self.sort_obj_iou:
j = iou.argsort()
b, a, gj, gi, iou = b[j], a[j], gj[j], gi[j], iou[j]通过gr用来设置IoU的值在objectness loss中做标签的比重。gr = 1 表示IOU是objectness的全部,gr < 1 表示iou的部分作为ogjectness
python
# 通过gr用来设置IoU的值在objectness loss中做标签的比重。
# gr = 1 表示IOU是objectness的全部,gr < 1 表示iou的部分作为ogjectness
if self.gr < 1:
iou = (1.0 - self.gr) + self.gr * iou注意下面这一部分不是按照代码的顺序,而是按照逻辑的顺序,用BCE loss完成损失值的计算。
pi[...,4] 是预测置信度,tboj是标注文件置信度,其中正样本用iou来填充,上面也详细说过,负样本都是0。
python
# 处理了正样本的置信度,用iou来做置信度。除了正样本之外,其他都是负样本,置信度都为0
tobj[b, a, gj, gi] = iou # iou ratio
obji = self.BCEobj(pi[..., 4], tobj)自动更新各个feature map的置信度损失系数
python
lobj += obji * self.balance[i] # obj loss
if self.autobalance:
self.balance[i] = self.balance[i] * 0.9999 + 0.0001 / obji.detach().item()类别损失
类别损失只有正样本参与计算。类别损失的正样本并不是0,而是通过平滑标签得到的一个近似1的值。
平滑标签介绍:
通常情况下,正确的边界框分类是用类的独热向量[0,0,0,1,0,0,...]来表示,并据此计算损失函数。
one-hot 编码存在的问题:
- 倾向于让模型更加"武断",成为一个"非黑即白"的模型,导致泛化性能差;
- 面对易混淆的分类任务、有噪音(误打标)的数据集时,更容易受影响。
根据这种直觉,对类标签表示进行编码以在某种程度上衡量不确定性更为合理。通常情况下,作者选择0.9,所以用[0,0,0,0.9,0...]来代表正确的类。
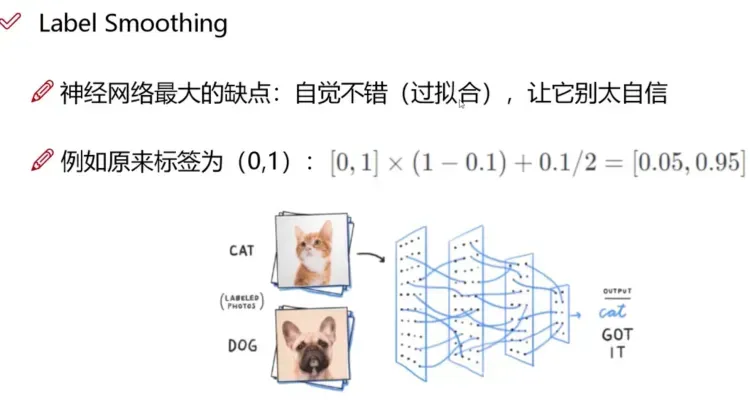
平滑标签的计算过程,默认情况下正样本:0.95, 负样本:0.05
python
self.cp, self.cn = smooth_BCE(eps=h.get('label_smoothing', 0.0)) # positive, negative BCE targets
def smooth_BCE(eps=0.1): # https://github.com/ultralytics/yolov3/issues/238#issuecomment-598028441
# return positive, negative label smoothing BCE targets
return 1.0 - 0.5 * eps, 0.5 * eps给正样本置信度赋值0.95之后,使用BCE loos计算函数,将预测值pcls和真实值t送入计算,得到结果lcls。
python
# Classification
if self.nc > 1: # cls loss (only if multiple classes)
# self.cn通过smooth_BCE平滑标签得到的,使得负样本不再是0,而是0.5 * eps
t = torch.full_like(pcls, self.cn, device=self.device) # targets
# self.cp 是通过smooth_BCE平滑标签得到的,使得正样本不再是1,而是1.0 - 0.5 * eps
t[range(n), tcls[i]] = self.cp
lcls += self.BCEcls(pcls, t) # BCE汇总损失值
根据超参中的损失权重参数 对各个损失进行平衡 防止总损失被某个损失所左右,
最后将3个损失值相加,并乘上bs,得到整个batch的总损失
python
lbox *= self.hyp['box']
lobj *= self.hyp['obj']
lcls *= self.hyp['cls']
bs = tobj.shape[0] # batch size
return (lbox + lobj + lcls) * bs, torch.cat((lbox, lobj, lcls)).detach()到这里损失函数的计算就完成了。
Q&A
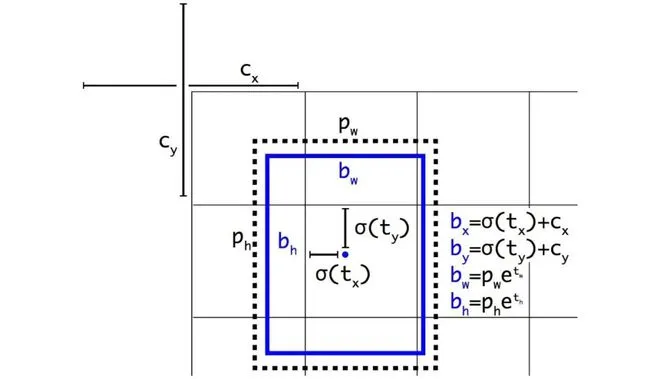
yolov5 bbox 相对于yolov3 计算方式的变化的原因是什么?
yolov3的边界框回归:
yolov5的边界框回归:
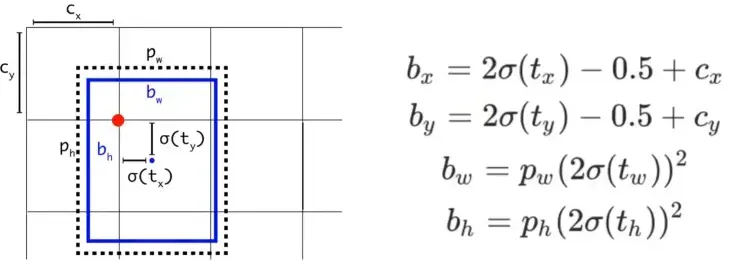
yolov5相对于yolov3主要改进有两个:
- 对中心点扩大范围,预测范围突破gt所在网格,扩展到左右0.5个网格
- 对宽高度加以限制,防止梯度失控。yolov3的e的n次方会呈指数上升,导致梯度过大。yolov5将宽高限制在0-4倍之间。
完整代码
附完整注释代码
python
def __call__(self, p, targets, imgs=None): # predictions, targets
lcls = torch.zeros(1, device=self.device) # class loss
lbox = torch.zeros(1, device=self.device) # box loss
lobj = torch.zeros(1, device=self.device) # object loss
tcls, tbox, indices, anchors = self.build_targets(p, targets, imgs) # targets
"""
tcls 保存类别id
tbox 保存的是gt中心相对于所在grid cell左上角偏移量。也会计算出gt中心相对扩展anchor的偏移量
indices 保存的内容是:image_id, anchor_id, grid x刻度 grid y刻度
anchors 保存anchor的具体宽高
"""
# Losses
"""
(Pdb) pp p[0].shape
torch.Size([1, 3, 80, 80, 7])
(Pdb) pp p[1].shape
torch.Size([1, 3, 40, 40, 7])
(Pdb) pp p[2].shape
torch.Size([1, 3, 20, 20, 7])
(Pdb) pp targets.sh
"""
for i, pi in enumerate(p): # layer index, layer predictions
b, a, gj, gi = indices[i] # image, anchor, gridy, gridx
tobj = torch.zeros(pi.shape[:4], dtype=pi.dtype, device=self.device) # target obj
n = b.shape[0] # number of targets
if n:
# pxy, pwh, _, pcls = pi[b, a, gj, gi].tensor_split((2, 4, 5), dim=1) # faster, requires torch 1.8.0
# 在这里就筛选出来了正样本相对应的预测结果
pxy, pwh, _, pcls = pi[b, a, gj, gi].split((2, 2, 1, self.nc), 1) # target-subset of predictions
# Regression
pxy = pxy.sigmoid() * 2 - 0.5
pwh = (pwh.sigmoid() * 2) ** 2 * anchors[i]
pbox = torch.cat((pxy, pwh), 1) # predicted box
"""
pbox 的xy相对于grid cell做了归一化处理 wh则是相对anchor所在feature map做的处理
同样 tbox 的xy也是相对grid cell 做的归一化处理,wh则没有做。
如何将同一个anchor下的box对应起来呢?通过筛选出来的正样本完成
"""
iou = bbox_iou(pbox, tbox[i], CIoU=True).squeeze() # iou(prediction, target)
lbox += (1.0 - iou).mean() # iou loss
# Objectness
iou = iou.detach().clamp(0).type(tobj.dtype)
if self.sort_obj_iou:
j = iou.argsort()
b, a, gj, gi, iou = b[j], a[j], gj[j], gi[j], iou[j]
# 通过gr用来设置IoU的值在objectness loss中做标签的比重。
# gr = 1 表示IOU是objectness的全部,gr < 1 表示iou的部分作为objectness
if self.gr < 1:
iou = (1.0 - self.gr) + self.gr * iou
# 处理了正样本的置信度,用iou来做置信度
tobj[b, a, gj, gi] = iou # iou ratio
# Classification
if self.nc > 1: # cls loss (only if multiple classes)
# self.cn通过smooth_BCE平滑标签得到的,使得负样本不再是0,而是0.5 * eps
t = torch.full_like(pcls, self.cn, device=self.device) # targets
# self.cp 是通过smooth_BCE平滑标签得到的,使得正样本不再是1,而是1.0 - 0.5 * eps
t[range(n), tcls[i]] = self.cp
lcls += self.BCEcls(pcls, t) # BCE
# Append targets to text file
# with open('targets.txt', 'a') as file:
# [file.write('%11.5g ' * 4 % tuple(x) + '\n') for x in torch.cat((txy[i], twh[i]), 1)]
obji = self.BCEobj(pi[..., 4], tobj)
lobj += obji * self.balance[i] # obj loss
if self.autobalance:
self.balance[i] = self.balance[i] * 0.9999 + 0.0001 / obji.detach().item()
if self.autobalance:
self.balance = [x / self.balance[self.ssi] for x in self.balance]
lbox *= self.hyp['box']
lobj *= self.hyp['obj']
lcls *= self.hyp['cls']
bs = tobj.shape[0] # batch size
return (lbox + lobj + lcls) * bs, torch.cat((lbox, lobj, lcls)).detach()