基于BiLSTM+CRF实现NER
学习目标
- 理解BiLSTM实现NER的原理
- 理解什么是CRF算法
- 掌握CRF的损失函数原理
1 BiLSTM+CRF模型介绍
-
BiLSTM+CRF模型的作用
propertiesBiLSTM+CRF: 解决NER问题 实现方式: 从一段自然语言文本中找出相关实体,并标注出其位置以及类型。 命名实体识别问题实际上是序列标注问题: 以中文分词任务进行举例, 例如输入序列是一串文字: "我是中国人", 输出序列是一串标签: "OOBII", 其中"BIO"组成了一种中文分词的标签体系: B表示这个字是词的开始, I表示词的中间到结尾, O表示其他类型词. 因此我们可以根据输出序列"OOBII"进行解码, 得到分词结果"我\是\中国人"。 序列标注问题涵盖了自然语言处理中的很多任务:包括语音识别, 中文分词, 机器翻译, 命名实体识别等, 而常见的序列标注模型包括HMM, CRF, RNN, LSTM, GRU等模型。 其中在命名实体识别技术上, 目前主流的技术: 通过BiLSTM+CRF模型进行序列标注。 -
模型架构(图1)
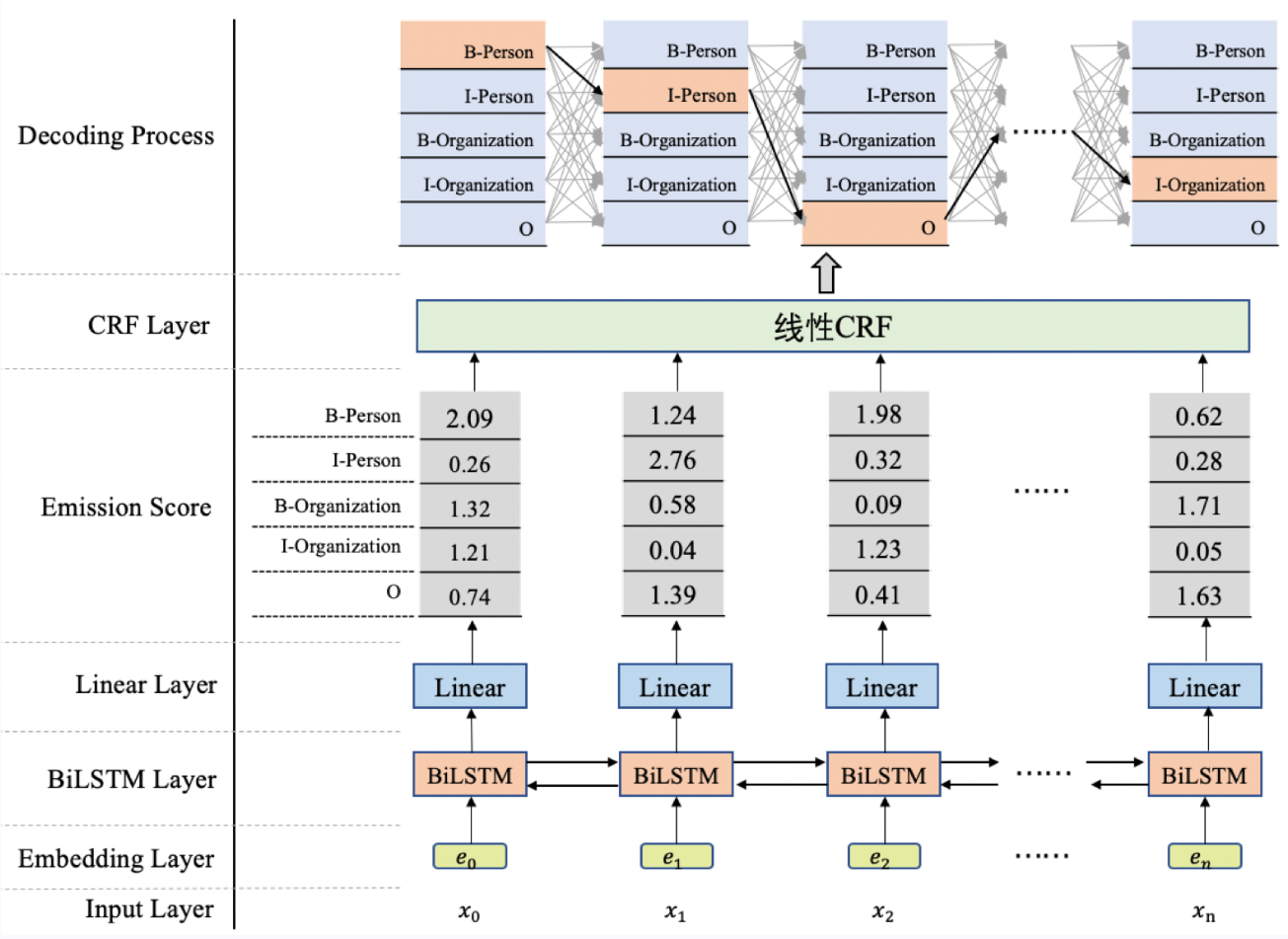
- 使用BiLSTM+CRF模型架构实现NER任务,大致分为两个阶段:
- 使用BiLSTM生成发射分数(标签向量)。
- 基于发射分数使用CRF解码最优的标签路径。
2 CRF原理
-
CRF在NER任务中的作用:
- 可以对标签序列之间起到约束左右
- 核心特征:转移概率矩阵和发射概率矩阵(来自于BiLSTM模型)
-
CRF建模的损失函数
- 简单概括:真实路径分数/所有路径分数之和
- 具体公式:
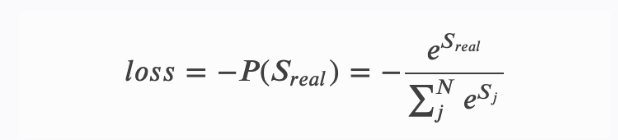
-
这个损失函数包含两部分:
- 单条真实路径的分数𝑆𝑟𝑒𝑎𝑙和归一化项𝑙𝑜𝑔(𝑒𝑆1+𝑒𝑆2+...+𝑒𝑆𝑁),
- 即将全部的路径分数进行𝑙𝑜𝑔_𝑠𝑢𝑚_𝑒𝑥𝑝操作,即先将每条路径分数𝑆𝑖进行𝑒𝑥𝑝(𝑆𝑖),然后再将所有的项加起来,最后取𝑙𝑜𝑔值。
-
注意:
- 使用前向算法计算所有路径分数。
- 前向算法和穷举所有路径的算法在时间复杂度上的差异显著:假设序列长度为 n,每个位置可能有 k 个标签:
- 穷举法:时间复杂度为 O(k^n)
- 前向算法:时间复杂度为 O(n* k^2),
- 使用维特比算法预测最优路径
- 其复杂度和前向算法一样
3 BiLSTM+CRF项目完整实现
基本步骤:
- 数据预处理
- 构建模型
- 模型训练和验证
- 模型预测
数据预处理
构造序列标注数据
-
需要对上述的origin数据进行处理,进行序列标注,选择标注方式:BIO,目的:对句子中的每个token都要标记上对应的标签,并统计所有tokens(去重后)并保存
举例说明:
- 未标注前:
text['以', '咳', '嗽', ',', '咳', '痰', ',', '发', '热', '为', '主', '症', '。']- 标注后:
text['O', 'B-SIGNS', 'I-SIGNS', 'O', 'B-SIGNS', 'I-SIGNS', 'O', 'B-SIGNS', 'I-SIGNS', 'O', 'O', 'O', 'O'] -
构造序列标注数据
- 代码路径:/MedicalKB/Ner/LSTM_CRF/utils/data_process.py
- 在data_process.py脚本中,定义一个TransferData类,实现数据格式的转换
- 具体代码:
pythonimport json import os from collections import Counter os.chdir('..') cur = os.getcwd() print('当前数据处理默认工作目录:', cur) class TransferData(): def __init__(self): self.label_dict = json.load(open(os.path.join(cur, 'data/labels.json'))) self.seq_tag_dict = json.load(open(os.path.join(cur,'data/tag2id.json'))) self.origin_path = os.path.join(cur, 'data_origin') self.train_filepath = os.path.join(cur, 'data/train.txt') def transfer(self): with open(self.train_filepath, 'w', encoding='utf-8') as fr: for root, dirs, files in os.walk(self.origin_path): for file in files: filepath = os.path.join(root, file) if 'original' not in filepath: continue label_filepath = filepath.replace('.txtoriginal','') print(filepath, '\t\t', label_filepath) res_dict = self.read_label_text(label_filepath) with open(filepath, 'r', encoding='utf-8')as f: content = f.read().strip() for indx, char in enumerate(content): char_label = res_dict.get(indx, 'O') fr.write(char + '\t' + char_label + '\n') def read_label_text(self, label_filepath): res_dict = {} for line in open(label_filepath, 'r', encoding='utf-8'): # line--》[右髋部\t21\t23\t身体部位] res = line.strip().split('\t') # res-->['右髋部', '21', '23', '身体部位'] start = int(res[1]) end = int(res[2]) label = res[3] label_tag = self.label_dict.get(label) for i in range(start, end + 1): if i == start: tag = "B-" + label_tag else: tag = "I-" + label_tag res_dict[i] = tag return res_dict if __name__ == '__main__': handler = TransferData() handler.transfer()经过数据转换后,我们会得到一个train.txt文档,此时,对于所有origin中的数据,都实现了BIO标注
text咳 B-SIGNS 痰 I-SIGNS , O 发 B-SIGNS 热 I-SIGNS , O 饮 O 食 O 、 O 睡 O 眠 O 尚 O 可 O
编写Config类项目文件配置代码
- Config类文件路径为:/MedicalKB/Ner/LSTM_CRF/config.py
- config文件目的: 配置项目常用变量,一般这些变量属于不经常改变的,比如: 训练文件路径、模型训练次数、模型超参数等等
python
import os
import torch
import json
class Config(object):
def __init__(self):
# 如果是windows或者linux电脑(使用GPU)
# self.device = "cuda:0" if torch.cuda.is_available() else "cpu:0"
# M1芯片及其以上的电脑(使用GPU)
self.device = 'mps'
self.train_path = '/MedicalKB/Ner/LSTM_CRF/data/train.txt'
self.vocab_path = '/MedicalKB/Ner/LSTM_CRF/vocab/vocab.txt'
self.embedding_dim = 300
self.epochs = 5
self.batch_size = 8
self.hidden_dim = 256
self.lr = 2e-3 # crf的时候,lr可以小点,比如1e-3
self.dropout = 0.2
self.model = "BiLSTM_CRF" # 可以只用"BiLSTM"
self.tag2id = json.load(open('/MedicalKB/Ner/LSTM_CRF/data/tag2id.json'))
if __name__ == '__main__':
conf = Config()
print(conf.train_path)
print(conf.tag2id)编写数据处理相关函数
- 构造数据预处理函数,分为两步骤:
- 1.构造样本x以及标签y数据对,以及获取vocabs
- 2.构造数据迭代器
构造(x,y)样本对,以及获取vocabs
- 因为在第二步构造序列标注数据时,没有对样本进行明确的分割,这里我们采用标点符号为分隔符,构造不同的(x, y)样本对
- 代码实现:
- 路径:/MedicalKB/Ner/LSTM_CRF/utils/common.py
python
from LSTM_CRF.config import *
conf = Config()
'''构造数据集'''
def build_data():
datas = []
sample_x = []
sample_y = []
vocab_list = ["PAD", 'UNK']
for line in open(conf.train_path, 'r', encoding='utf-8'):
line = line.rstrip().split('\t')
if not line:
continue
char = line[0]
if not char:
continue
cate = line[-1]
sample_x.append(char)
sample_y.append(cate)
if char not in vocab_list:
vocab_list.append(char)
if char in ['。', '?', '!', '!', '?']:
datas.append([sample_x, sample_y])
sample_x = []
sample_y = []
word2id = {wd: index for index, wd in enumerate(vocab_list)}
write_file(vocab_list, conf.vocab_path)
return datas, word2id
'''保存字典文件'''
def write_file(wordlist, filepath):
with open(filepath, 'w', encoding='utf-8') as f:
f.write('\n'.join(wordlist))
if __name__ == '__main__':
datas, word2id = build_data()
print(len(datas))
print(datas[:4])
print(word2id)
print(len(word2id))构造数据迭代器
-
代码路径:/MedicalKB/Ner/LSTM_CRF/utils/data_loader.py
-
第一步:导入必备的工具包
pythonimport json import torch from common import * from torch.utils.data import DataLoader, Dataset from torch.nn.utils.rnn import pad_sequence datas, word2id = build_data() -
第二步:构建Dataset类
python
class NerDataset(Dataset):
def __init__(self, datas):
super().__init__()
self.datas = datas
def __len__(self):
return len(self.datas)
def __getitem__(self, item):
x = self.datas[item][0]
y = self.datas[item][1]
return x, y- 第三步:构建自定义函数collate_fn()
python
def collate_fn(batch):
x_train = [torch.tensor([word2id[char] for char in data[0]]) for data in batch]
y_train = [torch.tensor([conf.tag2id[label] for label in data[1]]) for data in batch]
# 补齐input_ids, 使用0作为填充值
input_ids_padded = pad_sequence(x_train, batch_first=True, padding_value=0)
# # 补齐labels,注意如果使用了CRF,用0补齐,如果不用CRF,用-100补齐
labels_padded = pad_sequence(y_train, batch_first=True, padding_value=-100)
# 创建attention mask
attention_mask = (input_ids_padded != 0).long()
return input_ids_padded, labels_padded, attention_mask- 第四步:构建get_data函数,获得数据迭代器
python
def get_data():
train_dataset = NerDataset(datas[:6200])
train_dataloader = DataLoader(dataset=train_dataset,
batch_size=conf.batch_size,
collate_fn=collate_fn,
drop_last=True,
)
dev_dataset = NerDataset(datas[6200:])
dev_dataloader = DataLoader(dataset=dev_dataset,
batch_size=conf.batch_size,
collate_fn=collate_fn,
drop_last=True,
)
return train_dataloader, dev_dataloader
if __name__ == '__main__':
train_dataloader, dev_dataloader = get_data()
for input_ids_padded, labels_padded, attention_mask in train_dataloader:
print(input_ids_padded.shape)
print(labels_padded.shape)
print(attention_mask.shape)
breakBiLSTM+CRF模型搭建
- 构建BiLSTM模型
- 代码路径: /MedicalKB/Ner/LSTM_CRF/model/BiLSTM.py
python
import torch
import torch.nn as nn
class NERLSTM(nn.Module):
def __init__(self, embedding_dim, hidden_dim, dropout, word2id, tag2id):
super(NERLSTM, self).__init__()
self.name = "BiLSTM"
self.embedding_dim = embedding_dim
self.hidden_dim = hidden_dim
self.vocab_size = len(word2id) + 1
self.tag_to_ix = tag2id
self.tag_size = len(tag2id)
self.word_embeds = nn.Embedding(self.vocab_size, self.embedding_dim)
self.dropout = nn.Dropout(dropout)
self.lstm = nn.LSTM(self.embedding_dim, self.hidden_dim // 2,
bidirectional=True, batch_first=True)
self.hidden2tag = nn.Linear(self.hidden_dim, self.tag_size)
def forward(self, x, mask):
embedding = self.word_embeds(x)
outputs, hidden = self.lstm(embedding)
outputs = outputs * mask.unsqueeze(-1) # 仅保留有效位置的输出
outputs = self.dropout(outputs)
outputs = self.hidden2tag(outputs)
return outputs- 构建BiLSTM_CRF模型类
- 代码路径: /MedicalKB/Ner/LSTM_CRF/model/BiLSTM_CRF.py
python
import torch
import torch.nn as nn
from TorchCRF import CRF
class NERLSTM_CRF(nn.Module):
def __init__(self, embedding_dim, hidden_dim, dropout, word2id, tag2id):
super(NERLSTM_CRF, self).__init__()
self.name = "BiLSTM_CRF"
self.embedding_dim = embedding_dim
self.hidden_dim = hidden_dim
self.vocab_size = len(word2id) + 1
self.tag_to_ix = tag2id
self.tag_size = len(tag2id)
self.word_embeds = nn.Embedding(self.vocab_size, self.embedding_dim)
self.dropout = nn.Dropout(dropout)
#CRF
self.lstm = nn.LSTM(self.embedding_dim, self.hidden_dim // 2,
bidirectional=True, batch_first=True)
self.hidden2tag = nn.Linear(self.hidden_dim, self.tag_size)
self.crf = CRF(self.tag_size)
def forward(self, x, mask):
#lstm模型得到的结果
outputs = self.get_lstm2linear(x)
outputs = outputs * mask.unsqueeze(-1)
outputs = self.crf.viterbi_decode(outputs, mask)
return outputs
def log_likelihood(self, x, tags, mask):
# lstm模型得到的结果
outputs = self.get_lstm2linear(x)
outputs = outputs * mask.unsqueeze(-1)
# 计算损失
return - self.crf(outputs, tags, mask)
def get_lstm2linear(self, x):
embedding = self.word_embeds(x)
outputs, hidden = self.lstm(embedding)
outputs = self.dropout(outputs)
outputs = self.hidden2tag(outputs)
return outputs编写训练函数
- 实现训练函数train.py
- 代码位置:/MedicalKB/Ner/LSTM_CRF/trian.py
- 第一步:导入必备工具包
python
import torch
import torch.nn as nn
import torch.optim as optim
from model.BiLSTM import *
from model.BiLSTM_CRF import *
from utils.data_loader import *
from tqdm import tqdm
# classification_report可以导出字典格式,修改参数:output_dict=True,可以将字典在保存为csv格式输出
from sklearn.metrics import precision_score, recall_score, f1_score, classification_report
from config import *
conf = Config()- 第二步:实现模型训练函数的搭建:mode2trian()
python
def model2train():
# 获取数据
train_dataloader, dev_dataloader = get_data()
# 实例化模型
models = {'BiLSTM': NERLSTM,
'BiLSTM_CRF': NERLSTM_CRF}
model = models[conf.model](conf.embedding_dim, conf.hidden_dim, conf.dropout, word2id, conf.tag2id)
model = model.to(conf.device)
# 实例化损失函数
criterion = nn.CrossEntropyLoss()
# 实例化优化器
optimizer = optim.Adam(model.parameters(), lr=conf.lr)
# 选择模型进行训练
start_time = time.time()
if conf.model == 'BiLSTM':
f1_score = -1000
for epoch in range(conf.epochs):
model.train()
for index, (inputs, labels, mask) in enumerate(tqdm(train_dataloader, desc='BiLSTM训练')):
x = inputs.to(conf.device)
mask = mask.to(conf.device)
y = labels.to(conf.device)
pred = model(x, mask)
pred = pred.view(-1, len(conf.tag2id))
my_loss = criterion(pred, y.view(-1))
optimizer.zero_grad()
my_loss.backward()
optimizer.step()
if index % 200 == 0:
print('epoch:%04d,------------loss:%f' % (epoch, my_loss.item()))
precision, recall, f1, report = model2dev(dev_dataloader, model, criterion)
if f1 > f1_score:
f1_score = f1
torch.save(model.state_dict(), 'save_model/bilstm_best.pth')
print(report)
end_time = time.time()
print(f'训练总耗时:{end_time - start_time}')
elif conf.model == 'BiLSTM_CRF':
f1_score = -1000
for epoch in range(conf.epochs):
model.train()
for index, (inputs, labels, mask)in enumerate(tqdm(train_dataloader, desc='bilstm+crf训练')):
x = inputs.to(conf.device)
mask = mask.to(torch.bool).to(conf.device)
tags = labels.to(conf.device)
# CRF
loss = model.log_likelihood(x, tags, mask).mean()
optimizer.zero_grad()
loss.backward()
# CRF
torch.nn.utils.clip_grad_norm_(parameters=model.parameters(), max_norm=10)
optimizer.step()
if index % 200 == 0:
print('epoch:%04d,------------loss:%f' % (epoch, loss.item()))
precision, recall, f1, report = model2dev(dev_dataloader, model)
if f1 > f1_score:
f1_score = f1
torch.save(model.state_dict(), 'save_model/bilstm_crf_best.pth')
print(report)
end_time = time.time()
print(f'训练总耗时:{end_time-start_time}')- 第三步:实现模型验证函数的搭建:model2dev()
python
def model2dev(dev_iter, model, criterion=None):
aver_loss = 0
preds, golds = [], []
model.eval()
for index, (inputs, labels, mask) in enumerate(tqdm(dev_iter, desc="测试集验证")):
val_x = inputs.to(conf.device)
mask = mask.to(conf.device)
val_y = labels.to(conf.device)
predict = []
if model.name == "BiLSTM":
pred = model(val_x, mask)
predict = torch.argmax(pred, dim=-1).tolist()
pred = pred.view(-1, len(conf.tag2id))
val_loss = criterion(pred, val_y.view(-1))
aver_loss += val_loss.item()
elif model.name == "BiLSTM_CRF":
mask = mask.to(torch.bool)
predict = model(val_x, mask)
loss = model.log_likelihood(val_x, val_y, mask)
aver_loss += loss.mean().item()
# 统计非0的,也就是真实标签的长度
leng = []
for i in val_y.cpu():
tmp = []
for j in i:
if j.item() > 0:
tmp.append(j.item())
leng.append(tmp)
# 提取真实长度的预测标签
for index, i in enumerate(predict):
preds.extend(i[:len(leng[index])])
# 提取真实长度的真实标签
for index, i in enumerate(val_y.tolist()):
golds.extend(i[:len(leng[index])])
aver_loss /= (len(dev_iter) * 64)
precision = precision_score(golds, preds, average='macro')
recall = recall_score(golds, preds, average='macro')
f1 = f1_score(golds, preds, average='macro')
report = classification_report(golds, preds)
return precision, recall, f1, report-
模型训练效果:
-
BiLSTM:训练5轮,总共耗时:105s
- BiLSTM+CRF:训练5轮,总共耗时:831s
- 整体对比:BiLSTM+CRF虽然比BiLSTM速度慢,但是精度有所提升,这个效果随着数据量的增加还会进一步提升。
编写模型预测函数
- 使用训练好的模型,随机抽取文本进行NER
- 代码位置: /MedicalKB/Ner/LSTM_CRF/ner_predict.py
- 第一步:导入必备的工具包
python
import torch.nn as nn
import torch.optim as optim
from model.BiLSTM import *
from model.BiLSTM_CRF import *
from utils.data_loader import *
from tqdm import tqdm
# 实例化模型
models = {'BiLSTM': NERLSTM,
'BiLSTM_CRF': NERLSTM_CRF}
model = models["BiLSTM_CRF"](conf.embedding_dim, conf.hidden_dim, conf.dropout, word2id, conf.tag2id)
model.load_state_dict(torch.load('save_model/bilstm_crf_best.pth'))
id2tag = {value: key for key, value in conf.tag2id.items()}- 第二步:实现模型预测函数:model2test
python
def model2test(sample):
x = []
for char in sample:
if char not in word2id:
char = "UNK"
x.append(word2id[char])
x_train = torch.tensor([x])
mask = (x_train != 0).long()
model.eval()
with torch.no_grad():
if model.name =="BiLSTM":
outputs = model(x_train, mask)
preds_ids = torch.argmax(outputs,dim=-1)[0]
tags = [id2tag[i.item()] for i in preds_ids]
else:
preds_ids = model(x_train, mask)
tags = [id2tag[i] for i in preds_ids[0]]
chars = [i for i in sample]
assert len(chars) == len(tags)
result = extract_entities(chars, tags)
return result
if __name__ == '__main__':
result = model2test(sample='小明的父亲患有冠心病及糖尿病,无手术外伤史及药物过敏史')
print(result)- 第三步:实现实体解析函数:extract_entities
python
def extract_entities(tokens, labels):
entities = []
entity = []
entity_type = None
for token, label in zip(tokens, labels):
if label.startswith("B-"): # 实体的开始
if entity: # 如果已经有实体,先保存
entities.append((entity_type, ''.join(entity)))
entity = []
entity_type = label.split('-')[1]
entity.append(token)
elif label.startswith("I-") and entity: # 实体的中间或结尾
entity.append(token)
else:
if entity: # 保存上一个实体
entities.append((entity_type, ''.join(entity)))
entity = []
entity_type = None
# 如果最后一个实体没有保存,手动保存
if entity:
entities.append((entity_type, ''.join(entity)))
return {entity: entity_type for entity_type, entity in entities}