在深度学习模型中,McNemar检验(McNemar's Test,麦克尼马尔检验)是检验不同的深度学习分类器模型(准确率)或回归模型(命中率)在相同测试数据上性能是否存在显著差异的统计检验方法。
McNemar检验基于两个模型预测结果的2x2列联表(contingency table),如下图所示:

-
列联表:是对两个分类或回归变量进行列表或计数。列联表的依据是两个分类模型或回归模型均使用完全相同的训练数据进行训练,并使用完全相同的测试数据进行评估。
-
McNemar检验仅关注两个条件存在差异的单元格:McNemar仅关心n01和n10。n00:模型A和模型B均未命中的样本数。n11:模型A和模型B均命中的样本数。n01:模型A未命中,但模型B命中的样本数。n10:模型A命中,但模型B未命中的样本数。
-
判断逻辑:如果n01约等于n10:两模型差异可能是随机的。如果n01 >> n10:模型B显著优于模型A。如果n01 << n10:模型A显著优于模型B。
-
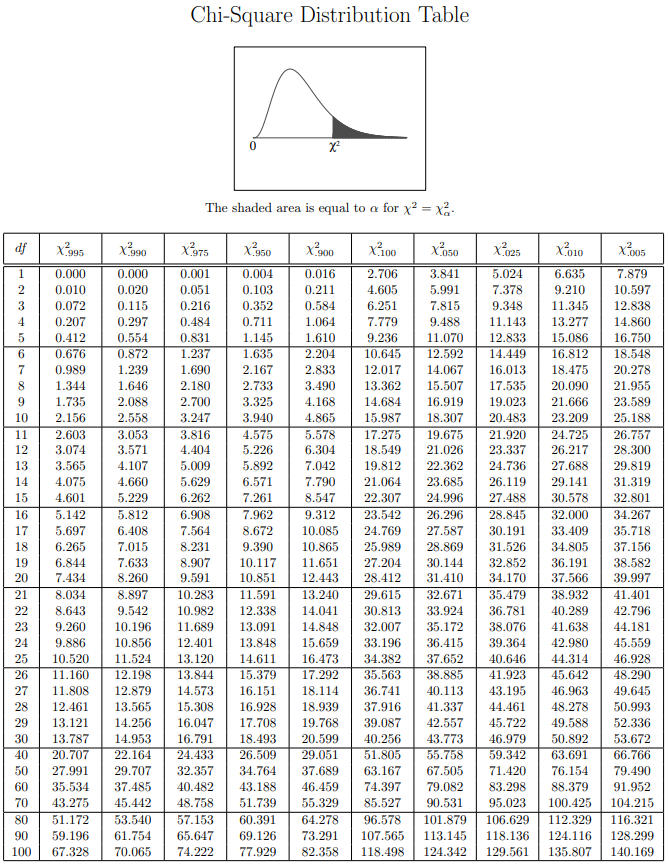
卡方分布表(chi-square distribution table),如下图所示:
- 公式:推荐使用连续性校正
(1).原始公式(未校正):10 <= n01 + n10 < 25,服从自由度df=1(或χ²(1))的卡方分布
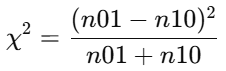
(2).连续性校正(Yates)公式:n01 + n10 >= 25,服从自由度df=1(或χ²(1))的卡方分布
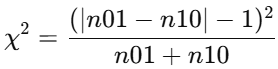
(3).精确检验:n01 + n10 < 10,scipy.stats.binomtest
- 判断标准,计算pvalue得结论:
(1).pvalue < 0.05:两个模型有显著差异。
(2).pvalue >= 0.05:两个模型差异不显著。
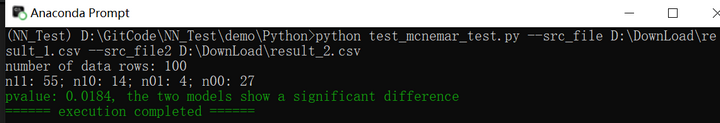
以下测试代码是用McNemar检验两个回归模型是否有显著差异:
python
def parse_args():
parser = argparse.ArgumentParser(description="mcnemar test")
parser.add_argument("--src_file", required=True, type=str, help="src file name")
parser.add_argument("--src_file2", required=True, type=str, help="src file name")
parser.add_argument("--threshold", type=float, default=0.5, help="error margin")
args = parser.parse_args()
return args
def mcnemar_test(src_file, src_file2, threshold):
if src_file is None or not src_file or not Path(src_file).is_file():
raise ValueError(colorama.Fore.RED + f"{src_file} is not a file")
if src_file2 is None or not src_file2 or not Path(src_file2).is_file():
raise ValueError(colorama.Fore.RED + f"{src_file2} is not a file")
def parse_csv(file):
with open(file, "r", encoding="utf-8") as f:
reader = csv.reader(f)
all_rows = list(reader)
data = all_rows[1:-1] # remove the first and last rows
return data
data1 = parse_csv(src_file)
data2 = parse_csv(src_file2)
if len(data1) != len(data2):
raise ValueError(colorama.Fore.RED + f"length mismath: {src_file}:{len(data1)}, {src_file2}:{len(data2)}")
print(f"number of data rows: {len(data1)}")
is_same = all(row1[0] == row2[0] for row1, row2 in zip(data1, data2))
if not is_same:
raise ValueError(colorama.Fore.RED + f"image name mismatch: {src_file}, {src_file2}")
n11 = 0; n10 = 0; n01 = 0; n00 = 0
for i in range(len(data1)):
value1 = abs(float(data1[i][1]) - float(data1[i][2]))
value2 = abs(float(data2[i][1]) - float(data2[i][2]))
if value1 <= threshold and value2 <= threshold:
n11 += 1
elif value1 > threshold and value2 > threshold:
n00 += 1
elif value1 <= threshold and value2 > threshold:
n10 += 1
elif value1 > threshold and value2 <= threshold:
n01 += 1
else:
raise ValueError(colorama.Fore.RED + f"unsupported conditions: value: {value1}, {value2}")
print(f"n11: {n11}; n10: {n10}; n01: {n01}; n00: {n00}")
if n10 + n01 == 0:
print(colorama.Fore.YELLOW + "unable to test differences")
return
def calculate_pvalue(n10, n01, method): # method: 0:Yates; 1:original; 2:exact binomial test
if method == 0:
stat = (abs(n01 - n10) - 1) ** 2 / (n10 + n01)
return chi2.sf(stat, df=1)
elif method == 1:
stat = (n01 - n10) ** 2 / (n10 + n01)
return chi2.sf(stat, df=1)
else:
return binomtest(k=min(n10, n01), n=n10+n01, p=0.5, alternative="two-sided").pvalue
if n10 + n01 >= 25:
pvalue = calculate_pvalue(n10, n01, 0)
elif 10 <= n10 + n01 < 25:
pvalue = calculate_pvalue(n10, n01, 1)
else:
pvalue = calculate_pvalue(n10, n01, 2)
if pvalue < 0.05:
print(colorama.Fore.GREEN + f"pvalue: {pvalue:.4f}, the two models show a significant difference")
else:
print(colorama.Fore.YELLOW + f"pvalue: {pvalue:.4f}, the two models no not show a significant difference")
if __name__ == "__main__":
colorama.init(autoreset=True)
args = parse_args()
mcnemar_test(args.src_file, args.src_file2, args.threshold)
print(colorama.Fore.GREEN + "====== execution completed ======")执行结果如下图所示: