作者,Evil Genius
今天我们拉群,2025番外--linux、R、python培训 https://mp.weixin.qq.com/s/8E1vYJMNhe5m0DXieBHfzA?payreadticket=HM3XzM3dAiSdVyi83oiJ3UWd-zPIOKO1EMMnYwy9ASu_scWj_WEje8Xwiwq_z8JvgcLDfd0&scene=1&click_id=1, 这是所有将来从事生物信息行业的第一课,也是现在入职诺禾、华大等上市公司的必备技能。
https://mp.weixin.qq.com/s/8E1vYJMNhe5m0DXieBHfzA?payreadticket=HM3XzM3dAiSdVyi83oiJ3UWd-zPIOKO1EMMnYwy9ASu_scWj_WEje8Xwiwq_z8JvgcLDfd0&scene=1&click_id=1, 这是所有将来从事生物信息行业的第一课,也是现在入职诺禾、华大等上市公司的必备技能。
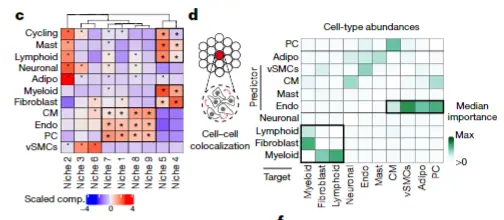
今天更新脚本,visium的细胞niche与共定位
单细胞空间联合推荐cell2location, 拿到联合后的结果,注意联合的时候尽量样本要匹配,联合关键的参数在课堂上都已经有详细的讲解。
我们来更新脚本
library(compositions)
library(tidyverse)
library(clustree)
library(uwot)
library(scran)
library(cluster)
integrated_compositions = read.csv('cell2location的联合矩阵')关键的一步,矩阵转换, 这个转换是等距对数比变换 (Isometric Log-Ratio Transformation),是处理比例、百分比等相对组成数据的专业方法。
其中ilr转换核心作用是解决成分数据的闭合性约束问题,使其能够在标准的多元统计方法中使用。
成分数据的根本问题
成分数据具有"闭合性"(总和为常数,如100%),这导致:
1、成分数据具有"闭合性"(总和为常数,如100%):伪相关性问题
2、非欧几里得几何:成分空间是单形空间,不是欧几里得空间,不能直接应用常规统计方法。
ilr转换的核心作用
1、消除闭合约束
2、保持等距性质
3. 实现坐标系统转换
# Generate ILR transformation
baseILR <- ilrBase(x = integrated_compositions,method = "basic")
cell_ilr <- as.matrix(ilr(integrated_compositions, baseILR))
colnames(cell_ilr) <- paste0("ILR_", 1:ncol(cell_ilr))
# Make community graph
k_vect <- c(10, 20,30)
k_vect <- set_names(k_vect, paste0("k_",k_vect))
cluster_info <- map(k_vect, function(k) {
print(k)
print("Generating SNN")
snn_graph <- scran::buildSNNGraph(x = t(cell_ilr %>% as.data.frame() %>% as.matrix()), k = k)
print("Louvain clustering")
clust.louvain <- igraph::cluster_louvain(snn_graph)
clusters <- tibble(cluster = clust.louvain$membership,
spot_id = rownames(cell_ilr))
})
cluster_info <- cluster_info %>%
enframe() %>%
unnest() %>%
pivot_wider(names_from = name,
values_from = cluster)
k_vect <- set_names(names(k_vect))
subsampling_mean_ss <- map(k_vect, function(k) {
print(k)
cluster_info_summary <- cluster_info %>%
group_by_at(k) %>%
summarize(ncells = floor(n() * 0.3))
cells <- cluster_info %>%
dplyr::select_at(c("spot_id", k)) %>%
group_by_at(k) %>%
nest() %>%
left_join(cluster_info_summary) %>%
mutate(data = map(data, ~ .x[[1]])) %>%
mutate(selected_cells = map2(data, ncells, function(dat,n) {
sample(dat, n)
})) %>%
pull(selected_cells) %>%
unlist()
dist_mat <- dist(cell_ilr[cells, ])
k_vect <- purrr::set_names(cluster_info[[k]], cluster_info[["spot_id"]])[cells]
sil <- cluster::silhouette(x = k_vect, dist = dist_mat)
mean(sil[, 'sil_width'])
})
subsampling_mean_ss <- enframe(subsampling_mean_ss) %>%
unnest() %>%
dplyr::filter()
niche_resolution <- dplyr::filter(subsampling_mean_ss,
value == max(value)) %>%
pull(name)
plt <- comp_umap %>%
ggplot(aes(x = V1, y = V2,
color = as.character(k_50))) +
ggrastr::geom_point_rast(size = 0.1) +
theme_classic() +
xlab("UMAP1") +
ylab("UMAP2") +
guides(colour = guide_legend(override.aes = list(size=4)))
plot(plt)
cts <- set_names(colnames(integrated_compositions))
walk(cts, function(ct){
plot_df <- comp_umap %>%
mutate(ct_prop = log_comps[ , ct])
plt <- plot_df %>%
ggplot(aes(x = V1, y = V2,
color = ct_prop)) +
ggrastr::geom_point_rast(size = 0.07) +
theme_classic() +
ggtitle(ct) +
xlab("UMAP1") +
ylab("UMAP2")
plot(plt)
})
dev.off()
# Make the niche annotation meta-data
cluster_info <- comp_umap %>%
dplyr::select(c("row_id","k_50")) %>%
dplyr::rename("niche" = k_50) %>%
dplyr::mutate(ct_niche = paste0("niche_", niche))
niche_summary_pat <- integrated_compositions %>%
as.data.frame() %>%
rownames_to_column("row_id") %>%
pivot_longer(-row_id,values_to = "ct_prop",
names_to = "cell_type") %>%
left_join(cluster_info) %>%
mutate(orig.ident = strsplit(row_id, "[..]") %>%
map_chr(., ~ .x[1])) %>%
group_by(orig.ident, ct_niche, cell_type) %>%
summarize(median_ct_prop = median(ct_prop))
niche_summary <- niche_summary_pat %>%
ungroup() %>%
group_by(ct_niche, cell_type) %>%
summarise(patient_median_ct_prop = median(median_ct_prop))
# Data manipulation to have clustered data
niche_summary_mat <- niche_summary %>%
pivot_wider(values_from = patient_median_ct_prop,
names_from = cell_type, values_fill = 0) %>%
column_to_rownames("ct_niche") %>%
as.matrix()
niche_order <- hclust(dist(niche_summary_mat))
niche_order <- niche_order$labels[niche_order$order]
ct_order <- hclust(dist(t(niche_summary_mat)))
ct_order <- ct_order$labels[ct_order$order]
# Find characteristic cell types of each niche
# We have per patient the proportion of each cell-type in each niche
run_wilcox_up <- function(prop_data) {
prop_data_group <- prop_data[["ct_niche"]] %>%
unique() %>%
set_names()
map(prop_data_group, function(g) {
test_data <- prop_data %>%
mutate(test_group = ifelse(ct_niche == g,
"target", "rest")) %>%
mutate(test_group = factor(test_group,
levels = c("target", "rest")))
wilcox.test(median_ct_prop ~ test_group,
data = test_data,
alternative = "greater") %>%
broom::tidy()
}) %>% enframe("ct_niche") %>%
unnest()
}
wilcoxon_res <- niche_summary_pat %>%
ungroup() %>%
group_by(cell_type) %>%
nest() %>%
mutate(wres = map(data, run_wilcox_up)) %>%
dplyr::select(wres) %>%
unnest() %>%
ungroup() %>%
mutate(p_corr = p.adjust(p.value))
wilcoxon_res <- wilcoxon_res %>%
mutate(significant = ifelse(p_corr <= 0.15, "*", ""))
write.table(niche_summary_pat, file = "./results/niche_mapping/ct_niches/niche_summary_pat.txt",
col.names = T, row.names = F, quote = F, sep = "\t")
write.table(wilcoxon_res, file = "./results/niche_mapping/ct_niches/wilcoxon_res_cells_niches.txt",
col.names = T, row.names = F, quote = F, sep = "\t")
mean_ct_prop_plt <- niche_summary %>%
left_join(wilcoxon_res, by = c("ct_niche", "cell_type")) %>%
mutate(cell_type = factor(cell_type, levels = ct_order),
ct_niche = factor(ct_niche, levels = niche_order)) %>%
ungroup() %>%
group_by(cell_type) %>%
mutate(scaled_pat_median = (patient_median_ct_prop - mean(patient_median_ct_prop))/sd(patient_median_ct_prop)) %>%
ungroup() %>%
ggplot(aes(x = cell_type, y = ct_niche, fill = scaled_pat_median)) +
geom_tile() +
geom_text(aes(label = significant)) +
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0.5, size = 12),
legend.position = "bottom",
plot.margin = unit(c(0, 0, 0, 0), "cm"),
axis.text.y = element_text(size=12)) +
scale_fill_gradient(high = "#ffd89b", low = "#19547b")
# Finally describe the proportions of those niches in all the data
cluster_counts <- cluster_info %>%
dplyr::select_at(c("row_id", "ct_niche")) %>%
group_by(ct_niche) %>%
summarise(nspots = length(ct_niche)) %>%
mutate(prop_spots = nspots/sum(nspots))
write_csv(cluster_counts, file = "./results/niche_mapping/ct_niches/niche_prop_summary.csv")
barplts <- cluster_counts %>%
mutate(ct_niche = factor(ct_niche, levels = niche_order)) %>%
ggplot(aes(y = ct_niche, x = prop_spots)) +
geom_bar(stat = "identity") +
theme_classic() + ylab("") +
theme(axis.text.y = element_blank(),
plot.margin = unit(c(0, 0, 0, 0), "cm"),
axis.text.x = element_text(size=12))
niche_summary_plt <- cowplot::plot_grid(mean_ct_prop_plt, barplts, align = "hv", axis = "tb")
pdf("./results/niche_mapping/ct_niches/characteristic_ct_niches.pdf", height = 3, width = 6)
plot(niche_summary_plt)
dev.off()
pdf(file = "./results/niche_mapping/ct_niches/niche_summary_pat.pdf", height = 5, width = 8)
niche_summary_pat %>%
ggplot(aes(x = ct_niche, y = median_ct_prop)) +
geom_boxplot() +
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0.5)) +
facet_wrap(.~cell_type, ncol = 3,scales = "free_y")
dev.off()