最近在写一个项目,用了BioPython的Entrez模块,因为写得比较长,所以try-except加的比较多。
但是以前只知道print出来error,而print其实根本不够,所以这次突发奇想多加了个
python
import traceback
# 在try-except的except分支中加入
traceback.print_exc()内容是运行了我自己写的一个命令行工具里的一个子命令,做一次数据测试:
python
✅ NCBI API Key set successfully. Rate limit increased to 10 req/s.
Fetching papers for query: alphafold3 AND conformation AND ensemble
Now searching PubMed with query [alphafold3 AND conformation AND ensemble] at [2025-12-19 15:43:23] ...
found 12 related articles about [alphafold3 AND conformation AND ensemble] at [2025-12-19 15:43:24] ...
Fetching articles 1 to 12 at [2025-12-19 15:43:24] ...
-> Retrieved 12 Medline records and 12 Xml articles. Please check whether they equal and the efetch number here with esearch count.
Error parsing Medline record PMID 41249430: [PaperContributors() takes no arguments] at [2025-12-19 15:43:27] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 656, in parse_medline_record
contributors=PaperContributors(
~~~~~~~~~~~~~~~~~^
medline={
^^^^^^^^^
...<4 lines>...
}
^
),
^
TypeError: PaperContributors() takes no arguments
Error parsing Medline record PMID 41147497: [PaperContributors() takes no arguments] at [2025-12-19 15:43:27] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 656, in parse_medline_record
contributors=PaperContributors(
~~~~~~~~~~~~~~~~~^
medline={
^^^^^^^^^
...<4 lines>...
}
^
),
^
TypeError: PaperContributors() takes no arguments
Error parsing Medline record PMID 41014267: [PaperContributors() takes no arguments] at [2025-12-19 15:43:27] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 656, in parse_medline_record
contributors=PaperContributors(
~~~~~~~~~~~~~~~~~^
medline={
^^^^^^^^^
...<4 lines>...
}
^
),
^
TypeError: PaperContributors() takes no arguments
Error parsing Medline record PMID 40950168: [PaperContributors() takes no arguments] at [2025-12-19 15:43:27] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 656, in parse_medline_record
contributors=PaperContributors(
~~~~~~~~~~~~~~~~~^
medline={
^^^^^^^^^
...<4 lines>...
}
^
),
^
TypeError: PaperContributors() takes no arguments
Error parsing Medline record PMID 40938899: [PaperContributors() takes no arguments] at [2025-12-19 15:43:27] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 656, in parse_medline_record
contributors=PaperContributors(
~~~~~~~~~~~~~~~~~^
medline={
^^^^^^^^^
...<4 lines>...
}
^
),
^
TypeError: PaperContributors() takes no arguments
Error parsing Medline record PMID 40714407: [PaperContributors() takes no arguments] at [2025-12-19 15:43:27] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 656, in parse_medline_record
contributors=PaperContributors(
~~~~~~~~~~~~~~~~~^
medline={
^^^^^^^^^
...<4 lines>...
}
^
),
^
TypeError: PaperContributors() takes no arguments
Error parsing Medline record PMID 40549150: [PaperContributors() takes no arguments] at [2025-12-19 15:43:27] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 656, in parse_medline_record
contributors=PaperContributors(
~~~~~~~~~~~~~~~~~^
medline={
^^^^^^^^^
...<4 lines>...
}
^
),
^
TypeError: PaperContributors() takes no arguments
Error parsing Medline record PMID 40490178: [PaperContributors() takes no arguments] at [2025-12-19 15:43:27] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 656, in parse_medline_record
contributors=PaperContributors(
~~~~~~~~~~~~~~~~~^
medline={
^^^^^^^^^
...<4 lines>...
}
^
),
^
TypeError: PaperContributors() takes no arguments
Error parsing Medline record PMID 39574676: [PaperContributors() takes no arguments] at [2025-12-19 15:43:27] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 656, in parse_medline_record
contributors=PaperContributors(
~~~~~~~~~~~~~~~~~^
medline={
^^^^^^^^^
...<4 lines>...
}
^
),
^
TypeError: PaperContributors() takes no arguments
Error parsing Medline record PMID 39186607: [PaperContributors() takes no arguments] at [2025-12-19 15:43:27] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 656, in parse_medline_record
contributors=PaperContributors(
~~~~~~~~~~~~~~~~~^
medline={
^^^^^^^^^
...<4 lines>...
}
^
),
^
TypeError: PaperContributors() takes no arguments
Error parsing Medline record PMID 38996889: [PaperContributors() takes no arguments] at [2025-12-19 15:43:27] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 656, in parse_medline_record
contributors=PaperContributors(
~~~~~~~~~~~~~~~~~^
medline={
^^^^^^^^^
...<4 lines>...
}
^
),
^
TypeError: PaperContributors() takes no arguments
Error parsing Medline record PMID 38995731: [PaperContributors() takes no arguments] at [2025-12-19 15:43:27] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 656, in parse_medline_record
contributors=PaperContributors(
~~~~~~~~~~~~~~~~~^
medline={
^^^^^^^^^
...<4 lines>...
}
^
),
^
TypeError: PaperContributors() takes no arguments
Saving 0 papers to storage...
Done.很显然能够看出来是一个循环,我们只看第1个循环
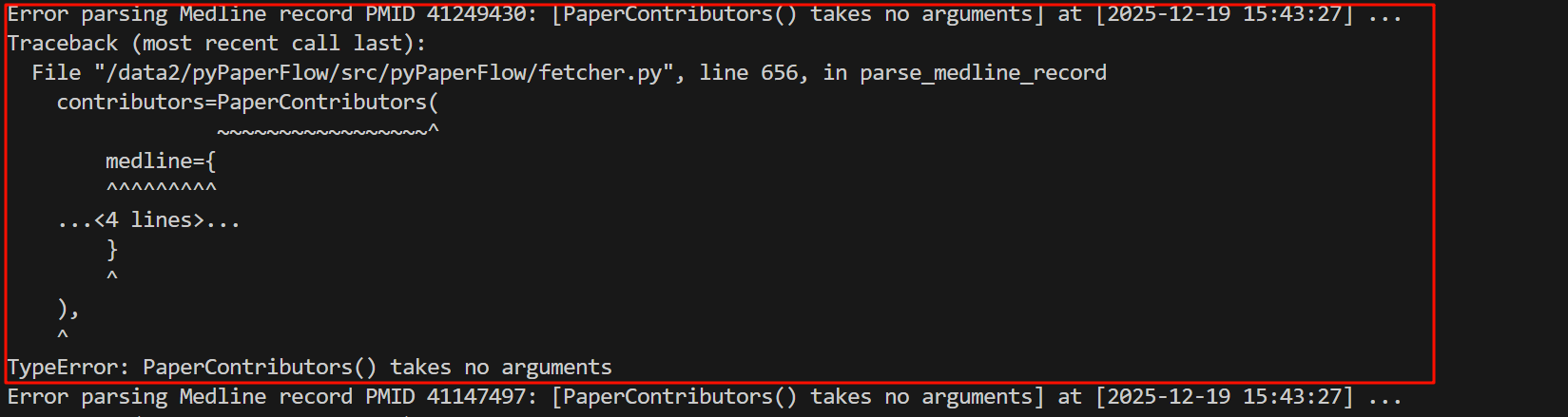
第1行是我自己的print提示不用看,就看下面的:Traceback~TypeError,
可以看到错误原因:也就是最下面的TypeError是PaperContributors这个类不接受参数初始化,
那么我们再回过头来看,具体报错的地方在fetcher.py这个脚本,在第656行,在parse_medline_record这个函数中。
错误的地方在于我们对这个类的medline属性进行了初始化赋值。
我们回到脚本中:
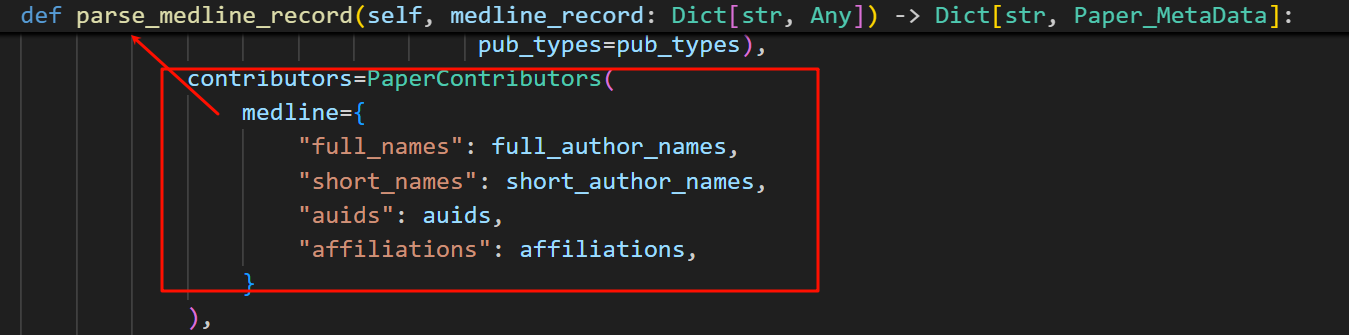
对于PaperContributors这个变量我们ctrl+左键,查看脚本中所有的引用
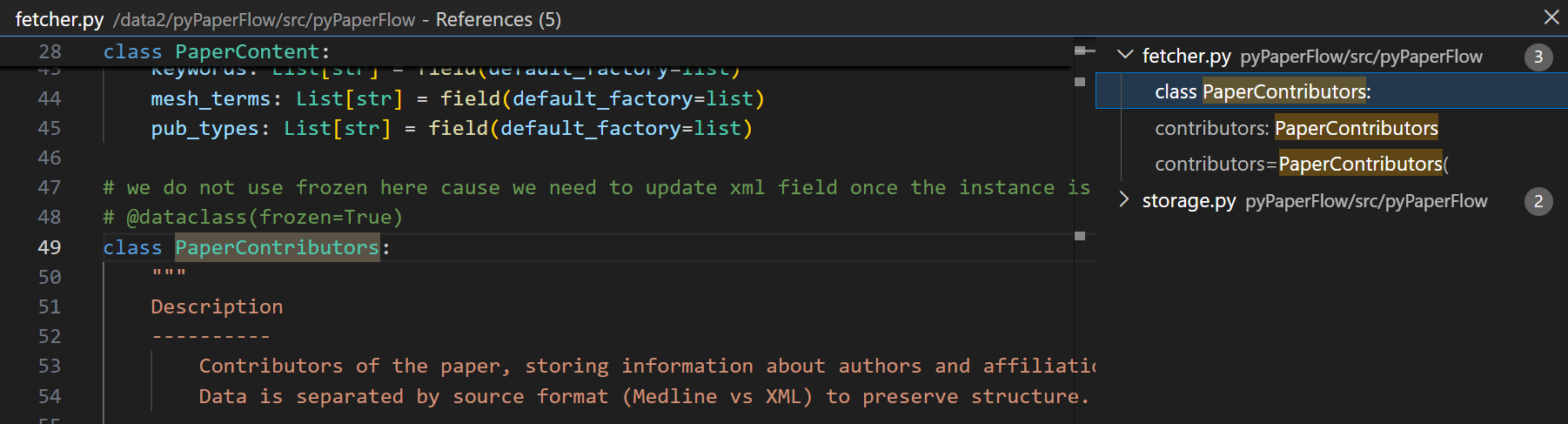
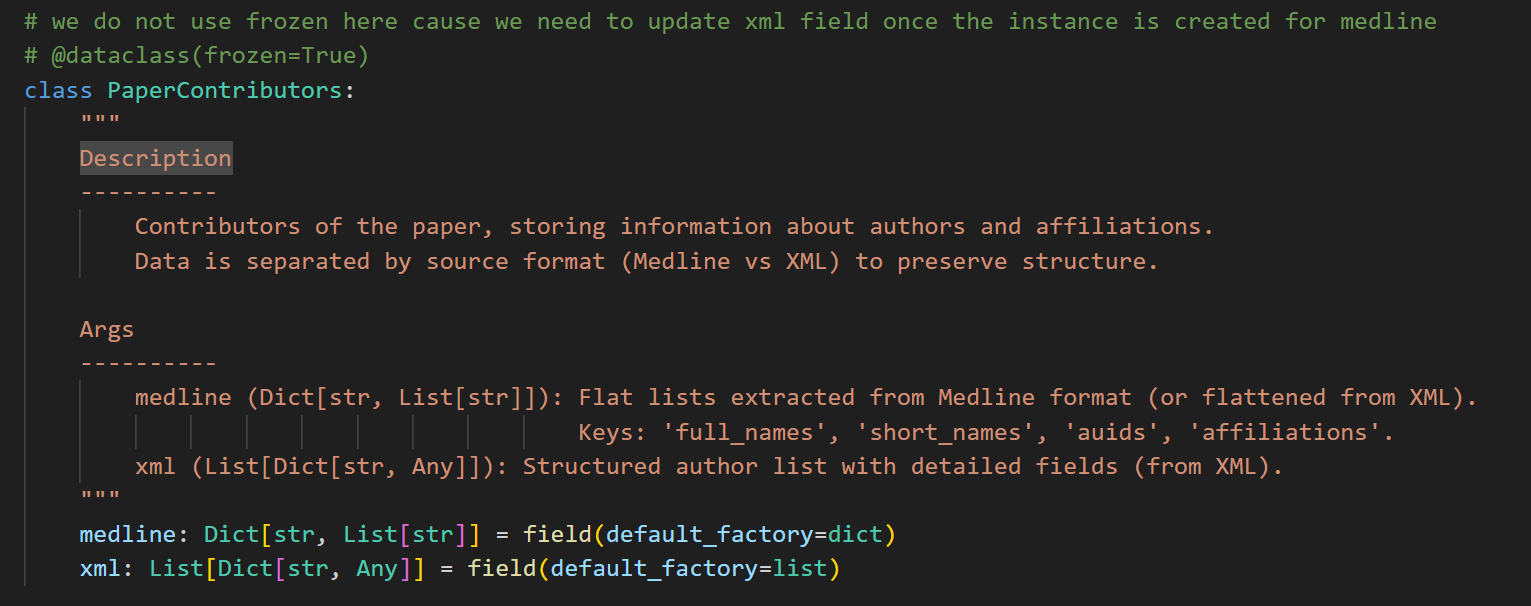
直接回到最开始的定义中:
可以发现我们这里是想写一个dataclass的,但是想去掉frozen=True这个限制。
这个时候我就想起来了,我为什么要注释掉这个限制:
- 首先我需要dataclass,因为不用写初始化等,比较轻便,具体可以参考我之前的博客:数据类dataclass
- 我写了两个解析函数,一个用于解析Medline格式的返回值,作为medline属性的值;另外一个用于解析XML格式的返回值,作为xml属性的值。但是我构建这个实例是先后调用这两个函数,第1次调用就是用medline数据实例化,第2次调用是更新xml数据,但是我写了frozen=True就没法更新数据了。因为我的逻辑是"先创建基础信息,再补充xml信息",所以这个类必须得是可变的,但是我写了frozen=True就意味着这个实例一旦创建,就不能够修改任何属性
所以我临时注释掉了,结果把dataclass也给注释掉了,结果就导致这个class不是dataclass,而是变成了一个普通的class,而普通的class是需要我们自己手写init方法的,不然默认的初始化函数是不接受任何参数的,所以这里就报错了。
然后简单修改了之后又报了另外一个错:
python
Error in batch Elink acheck for 12 PMIDs: [IncompleteRead(167 bytes read)] at [2025-12-19 16:27:02] ...
Traceback (most recent call last):
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/http/client.py", line 579, in _get_chunk_left
chunk_left = self._read_next_chunk_size()
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/http/client.py", line 546, in _read_next_chunk_size
return int(line, 16)
ValueError: invalid literal for int() with base 16: b''
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/http/client.py", line 597, in _read_chunked
while (chunk_left := self._get_chunk_left()) is not None:
~~~~~~~~~~~~~~~~~~~~^^
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/http/client.py", line 581, in _get_chunk_left
raise IncompleteRead(b'')
http.client.IncompleteRead: IncompleteRead(0 bytes read)
The above exception was the direct cause of the following exception:
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 950, in fetch_linked_data_for_batch_pmid
acheck_results = Entrez.read(handle)
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/site-packages/Bio/Entrez/__init__.py", line 529, in read
record = handler.read(source)
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/site-packages/Bio/Entrez/Parser.py", line 405, in read
self.parser.ParseFile(stream)
~~~~~~~~~~~~~~~~~~~~~^^^^^^^^
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/http/client.py", line 473, in read
return self._read_chunked(amt)
~~~~~~~~~~~~~~~~~~^^^^^
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/http/client.py", line 609, in _read_chunked
raise IncompleteRead(b''.join(value)) from exc
http.client.IncompleteRead: IncompleteRead(167 bytes read)
Fetching articles 1 to 12 failed: ['type' object does not support item assignment] at [2025-12-19 16:27:02] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 420, in fetch_from_query
links_map = self.fetch_linked_data_for_batch_pmid(current_batch_pmids) # Dict[str, PaperLinks]
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 960, in fetch_linked_data_for_batch_pmid
links_to_fetch = set[Tuple[str, str]] = set()
~~~^^^^^^^^^^^^^^^^^
TypeError: 'type' object does not support item assignment
Saving 0 papers to storage...
Done.同样的,我们按照下面单元来看
python
Traceback (most recent call last):
...
ErrorType: ErrorMessage注意到有direct cause,那么前面的不用看,基本上都是一些python内容的源码:
我们从这行下面的报错开始看:
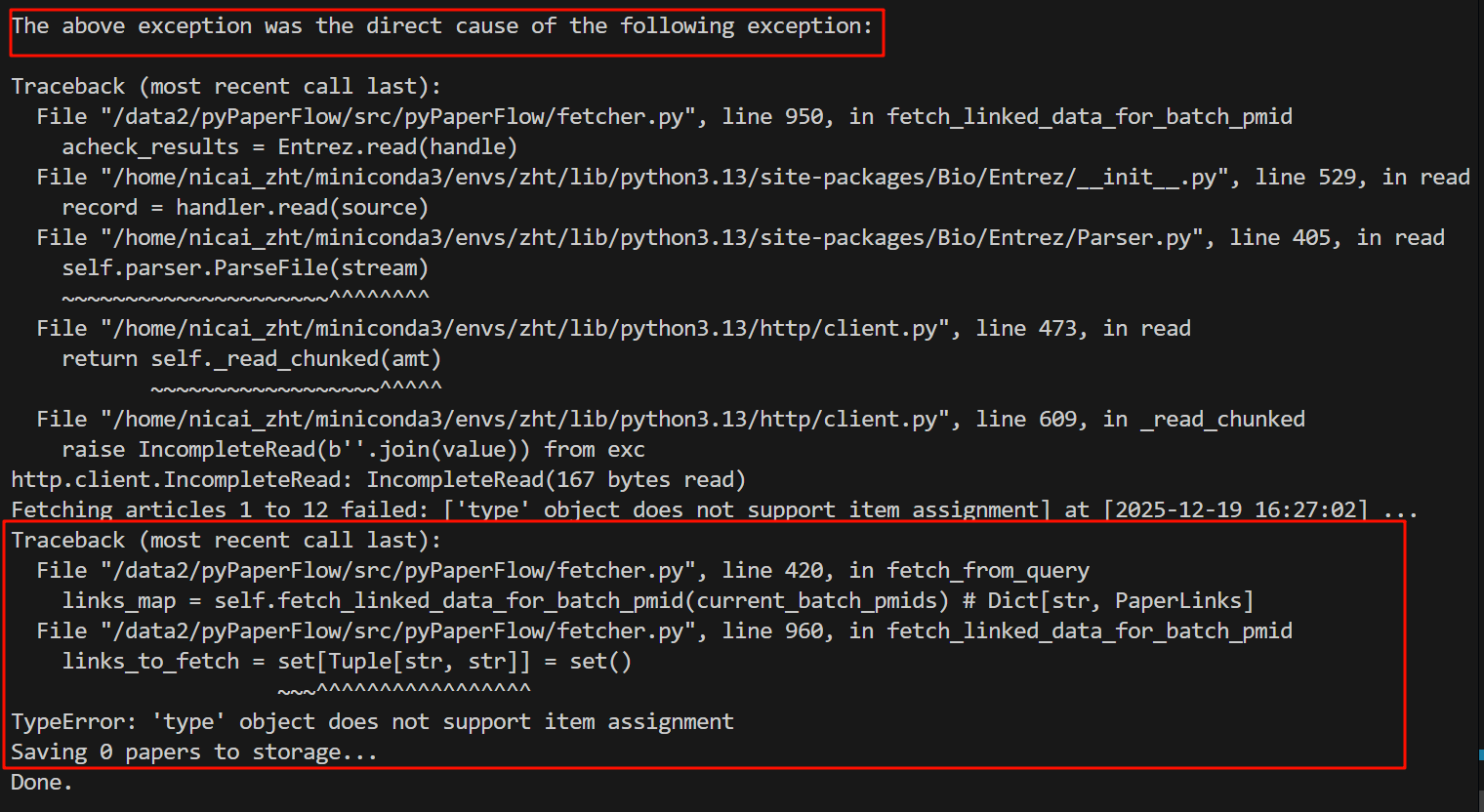
然后看的话都按照:most recent call last,也就是最近的调用在最下面

我们先看最后一个traceback,发现类型错误:
报错原因是因为类型对象不能给它赋值,我们正在尝试给一个类型对象赋值,
也就是右边的这个set[]=set()这一块,很显然我们左边写的这个是一个类型,集合类型,而集合类型的赋值没有这种操作方法,
所以能够看出来我这里其实是类型注解与默认赋值都写成了=号,第一个应该用冒号。
修改了之后报了第3个错
python
Error in batch Elink acheck for 12 PMIDs: [IncompleteRead(167 bytes read)] at [2025-12-19 16:49:50] ...
Traceback (most recent call last):
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/http/client.py", line 579, in _get_chunk_left
chunk_left = self._read_next_chunk_size()
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/http/client.py", line 546, in _read_next_chunk_size
return int(line, 16)
ValueError: invalid literal for int() with base 16: b''
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/http/client.py", line 597, in _read_chunked
while (chunk_left := self._get_chunk_left()) is not None:
~~~~~~~~~~~~~~~~~~~~^^
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/http/client.py", line 581, in _get_chunk_left
raise IncompleteRead(b'')
http.client.IncompleteRead: IncompleteRead(0 bytes read)
The above exception was the direct cause of the following exception:
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 950, in fetch_linked_data_for_batch_pmid
acheck_results = Entrez.read(handle)
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/site-packages/Bio/Entrez/__init__.py", line 529, in read
record = handler.read(source)
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/site-packages/Bio/Entrez/Parser.py", line 405, in read
self.parser.ParseFile(stream)
~~~~~~~~~~~~~~~~~~~~~^^^^^^^^
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/http/client.py", line 473, in read
return self._read_chunked(amt)
~~~~~~~~~~~~~~~~~~^^^^^
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/http/client.py", line 609, in _read_chunked
raise IncompleteRead(b''.join(value)) from exc
http.client.IncompleteRead: IncompleteRead(167 bytes read)
-> Deep mining 4 types of internal connections for 12 PMIDs at [2025-12-19 16:49:50] ...
Fetching pubmed_pubmed from pubmed for 12 PMIDs at [2025-12-19 16:49:50] ...
Fetching pubmed_pubmed_citedin from pubmed for 12 PMIDs at [2025-12-19 16:49:57] ...
Warning: Failed to parse fetched links for pubmed_pubmed_citedin from pubmed for 41014267: [list index out of range] at [2025-12-19 16:50:04] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 1026, in fetch_linked_data_for_batch_pmid
for uid in linkset['LinkSetDb'][0]['Link']:
~~~~~~~~~~~~~~~~~~~~^^^
IndexError: list index out of range
Warning: Failed to parse fetched links for pubmed_pubmed_citedin from pubmed for 40950168: [list index out of range] at [2025-12-19 16:50:04] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 1026, in fetch_linked_data_for_batch_pmid
for uid in linkset['LinkSetDb'][0]['Link']:
~~~~~~~~~~~~~~~~~~~~^^^
IndexError: list index out of range
Warning: Failed to parse fetched links for pubmed_pubmed_citedin from pubmed for 40938899: [list index out of range] at [2025-12-19 16:50:04] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 1026, in fetch_linked_data_for_batch_pmid
for uid in linkset['LinkSetDb'][0]['Link']:
~~~~~~~~~~~~~~~~~~~~^^^
IndexError: list index out of range
Warning: Failed to parse fetched links for pubmed_pubmed_citedin from pubmed for 40714407: [list index out of range] at [2025-12-19 16:50:04] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 1026, in fetch_linked_data_for_batch_pmid
for uid in linkset['LinkSetDb'][0]['Link']:
~~~~~~~~~~~~~~~~~~~~^^^
IndexError: list index out of range
Warning: Failed to parse fetched links for pubmed_pubmed_citedin from pubmed for 40549150: [list index out of range] at [2025-12-19 16:50:04] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 1026, in fetch_linked_data_for_batch_pmid
for uid in linkset['LinkSetDb'][0]['Link']:
~~~~~~~~~~~~~~~~~~~~^^^
IndexError: list index out of range
Warning: Failed to parse fetched links for pubmed_pubmed_citedin from pubmed for 39574676: [list index out of range] at [2025-12-19 16:50:04] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 1026, in fetch_linked_data_for_batch_pmid
for uid in linkset['LinkSetDb'][0]['Link']:
~~~~~~~~~~~~~~~~~~~~^^^
IndexError: list index out of range
Fetching pubmed_pubmed_reviews from pubmed for 12 PMIDs at [2025-12-19 16:50:04] ...
Fetching pubmed_pubmed_refs from pubmed for 12 PMIDs at [2025-12-19 16:50:11] ...
Warning: Failed to parse fetched links for pubmed_pubmed_refs from pubmed for 41147497: [list index out of range] at [2025-12-19 16:50:19] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 1026, in fetch_linked_data_for_batch_pmid
for uid in linkset['LinkSetDb'][0]['Link']:
~~~~~~~~~~~~~~~~~~~~^^^
IndexError: list index out of range
Warning: Failed to parse fetched links for pubmed_pubmed_refs from pubmed for 41014267: [list index out of range] at [2025-12-19 16:50:19] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 1026, in fetch_linked_data_for_batch_pmid
for uid in linkset['LinkSetDb'][0]['Link']:
~~~~~~~~~~~~~~~~~~~~^^^
IndexError: list index out of range
Warning: Failed to parse fetched links for pubmed_pubmed_refs from pubmed for 40714407: [list index out of range] at [2025-12-19 16:50:19] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 1026, in fetch_linked_data_for_batch_pmid
for uid in linkset['LinkSetDb'][0]['Link']:
~~~~~~~~~~~~~~~~~~~~^^^
IndexError: list index out of range
Warning: Failed to parse fetched links for pubmed_pubmed_refs from pubmed for 40490178: [list index out of range] at [2025-12-19 16:50:19] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 1026, in fetch_linked_data_for_batch_pmid
for uid in linkset['LinkSetDb'][0]['Link']:
~~~~~~~~~~~~~~~~~~~~^^^
IndexError: list index out of range
Warning: Failed to parse fetched links for pubmed_pubmed_refs from pubmed for 39574676: [list index out of range] at [2025-12-19 16:50:19] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 1026, in fetch_linked_data_for_batch_pmid
for uid in linkset['LinkSetDb'][0]['Link']:
~~~~~~~~~~~~~~~~~~~~^^^
IndexError: list index out of range
Warning: Failed to parse fetched links for pubmed_pubmed_refs from pubmed for 39186607: [list index out of range] at [2025-12-19 16:50:19] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 1026, in fetch_linked_data_for_batch_pmid
for uid in linkset['LinkSetDb'][0]['Link']:
~~~~~~~~~~~~~~~~~~~~^^^
IndexError: list index out of range
Warning: Failed to parse fetched links for pubmed_pubmed_refs from pubmed for 38996889: [list index out of range] at [2025-12-19 16:50:19] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 1026, in fetch_linked_data_for_batch_pmid
for uid in linkset['LinkSetDb'][0]['Link']:
~~~~~~~~~~~~~~~~~~~~^^^
IndexError: list index out of range
-> Fetching external LinkOuts (Datasets, Full Text, etc.) for 12 PMIDs at [2025-12-19 16:50:19] ...
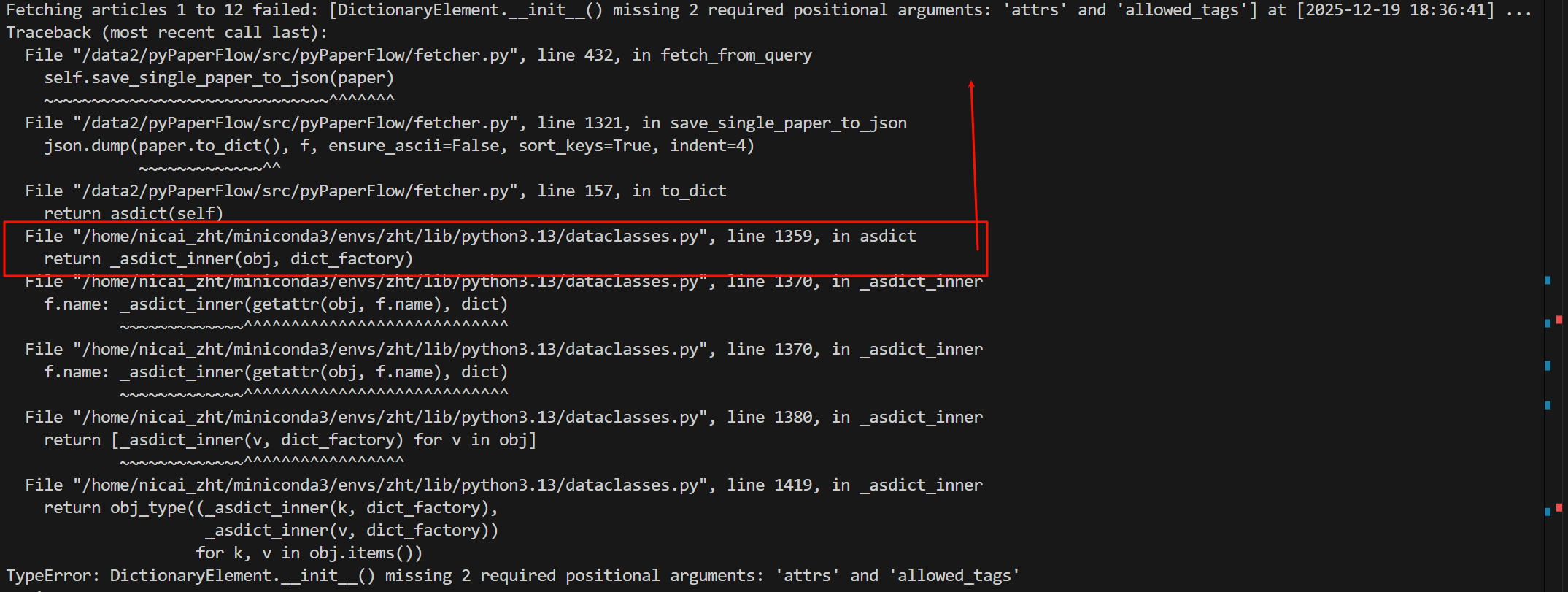
Fetching articles 1 to 12 failed: [DictionaryElement.__init__() missing 2 required positional arguments: 'attrs' and 'allowed_tags'] at [2025-12-19 16:50:23] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 432, in fetch_from_query
self.save_single_paper_to_json(paper)
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~^^^^^^^
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 1273, in save_single_paper_to_json
json.dump(paper.to_dict(), f, ensure_ascii=False, sort_keys=True, indent=4)
~~~~~~~~~~~~~^^
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 157, in to_dict
return asdict(self)
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/dataclasses.py", line 1359, in asdict
return _asdict_inner(obj, dict_factory)
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/dataclasses.py", line 1370, in _asdict_inner
f.name: _asdict_inner(getattr(obj, f.name), dict)
~~~~~~~~~~~~~^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/dataclasses.py", line 1370, in _asdict_inner
f.name: _asdict_inner(getattr(obj, f.name), dict)
~~~~~~~~~~~~~^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/dataclasses.py", line 1380, in _asdict_inner
return [_asdict_inner(v, dict_factory) for v in obj]
~~~~~~~~~~~~~^^^^^^^^^^^^^^^^^
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/dataclasses.py", line 1419, in _asdict_inner
return obj_type((_asdict_inner(k, dict_factory),
_asdict_inner(v, dict_factory))
for k, v in obj.items())
TypeError: DictionaryElement.__init__() missing 2 required positional arguments: 'attrs' and 'allowed_tags'
Saving 0 papers to storage...
Done.好在我在写try-except+堆栈调用前,做了比较完善的报错信息打印:


如图所示在direct cause下面我们看到了一个又一个的循环,
两套循环中都有地方报错,当然这是两个独立的过程,反正两个问题我们都要解决,先看第一个循环报错的地方,
然后循环里面我们先看第一个循环:
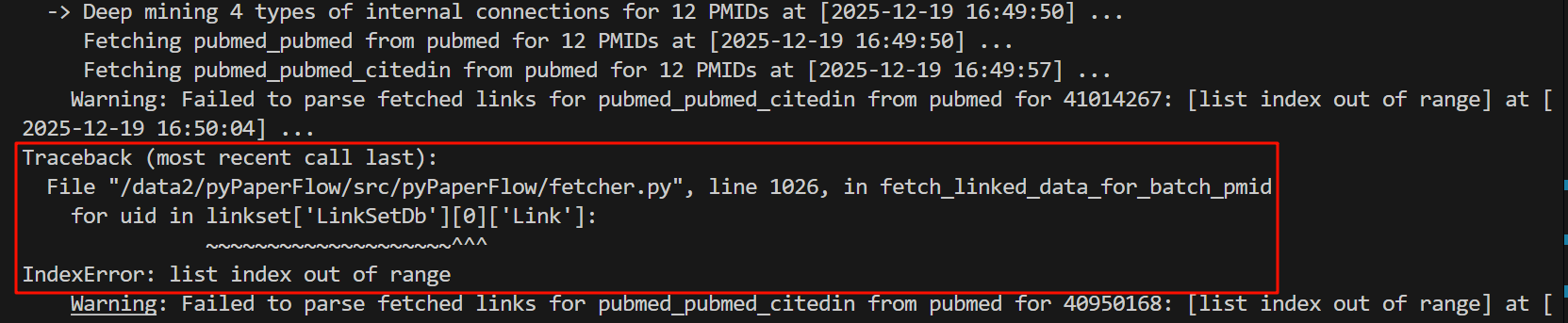
报错信息一目了然,就是下标越界,很常见的那种在迭代器中会犯的错误。
报错原因是因为我这里没有做防御性编程,虽然我已经多次循环continue。
一般的防御性编程:
对于字典,检查键值对是否存在,也就是用get方法而不是直接key取切片;
对于列表,索引越界,一般是检查查看再看看索引到哪里。
在简单对源代码修改了一点点防御性编程之后,再次运行,再次报错
python
Error in batch Elink acheck for 12 PMIDs: [IncompleteRead(167 bytes read)] at [2025-12-19 18:36:20] ...
Traceback (most recent call last):
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/http/client.py", line 579, in _get_chunk_left
chunk_left = self._read_next_chunk_size()
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/http/client.py", line 546, in _read_next_chunk_size
return int(line, 16)
ValueError: invalid literal for int() with base 16: b''
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/http/client.py", line 597, in _read_chunked
while (chunk_left := self._get_chunk_left()) is not None:
~~~~~~~~~~~~~~~~~~~~^^
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/http/client.py", line 581, in _get_chunk_left
raise IncompleteRead(b'')
http.client.IncompleteRead: IncompleteRead(0 bytes read)
The above exception was the direct cause of the following exception:
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 971, in fetch_linked_data_for_batch_pmid
acheck_results = Entrez.read(handle)
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/site-packages/Bio/Entrez/__init__.py", line 529, in read
record = handler.read(source)
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/site-packages/Bio/Entrez/Parser.py", line 405, in read
self.parser.ParseFile(stream)
~~~~~~~~~~~~~~~~~~~~~^^^^^^^^
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/http/client.py", line 473, in read
return self._read_chunked(amt)
~~~~~~~~~~~~~~~~~~^^^^^
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/http/client.py", line 609, in _read_chunked
raise IncompleteRead(b''.join(value)) from exc
http.client.IncompleteRead: IncompleteRead(167 bytes read)
-> Deep mining 4 types of internal connections for 12 PMIDs at [2025-12-19 18:36:20] ...
Fetching pubmed_pubmed from pubmed for 12 PMIDs at [2025-12-19 18:36:20] ...
Fetching pubmed_pubmed_citedin from pubmed for 12 PMIDs at [2025-12-19 18:36:27] ...
Fetching pubmed_pubmed_refs from pubmed for 12 PMIDs at [2025-12-19 18:36:32] ...
Fetching pubmed_pubmed_reviews from pubmed for 12 PMIDs at [2025-12-19 18:36:36] ...
-> Fetching external LinkOuts (Datasets, Full Text, etc.) for 12 PMIDs at [2025-12-19 18:36:39] ...
Fetching articles 1 to 12 failed: [DictionaryElement.__init__() missing 2 required positional arguments: 'attrs' and 'allowed_tags'] at [2025-12-19 18:36:41] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 432, in fetch_from_query
self.save_single_paper_to_json(paper)
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~^^^^^^^
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 1321, in save_single_paper_to_json
json.dump(paper.to_dict(), f, ensure_ascii=False, sort_keys=True, indent=4)
~~~~~~~~~~~~~^^
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 157, in to_dict
return asdict(self)
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/dataclasses.py", line 1359, in asdict
return _asdict_inner(obj, dict_factory)
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/dataclasses.py", line 1370, in _asdict_inner
f.name: _asdict_inner(getattr(obj, f.name), dict)
~~~~~~~~~~~~~^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/dataclasses.py", line 1370, in _asdict_inner
f.name: _asdict_inner(getattr(obj, f.name), dict)
~~~~~~~~~~~~~^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/dataclasses.py", line 1380, in _asdict_inner
return [_asdict_inner(v, dict_factory) for v in obj]
~~~~~~~~~~~~~^^^^^^^^^^^^^^^^^
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/dataclasses.py", line 1419, in _asdict_inner
return obj_type((_asdict_inner(k, dict_factory),
_asdict_inner(v, dict_factory))
for k, v in obj.items())
TypeError: DictionaryElement.__init__() missing 2 required positional arguments: 'attrs' and 'allowed_tags'
Saving 0 papers to storage...
Done.同样,direct cause前面不看,
我们直接看最后一个traceback,
同样是从最下面开始往上面看,看到我们熟悉的代码或者脚本为止,
比如说下面这里asdict就是我们脚本里调用的函数,往上面和我们的脚本相关了,就是第157行,我们写的一个to_dict函数
回到我们的原始脚本:
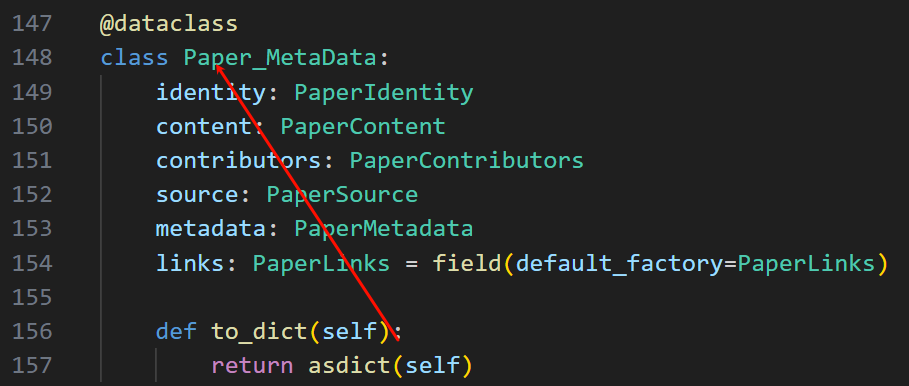
可以看到to_dict这个函数是写给paper_metadata类的,
没什么毛病,再往前看:
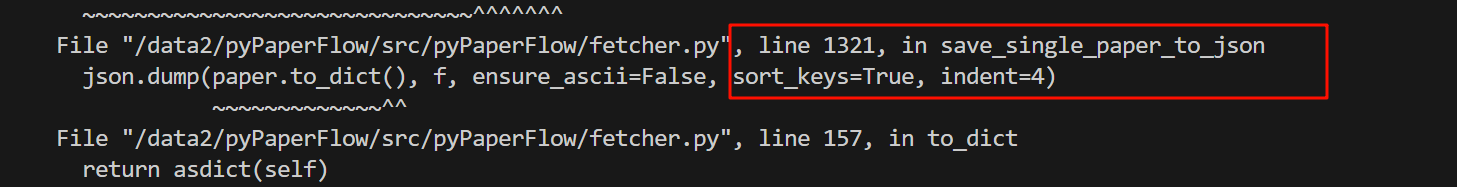
在save_single_paper_to_json这个函数中,在第1321行,调用了to_dict这个函数
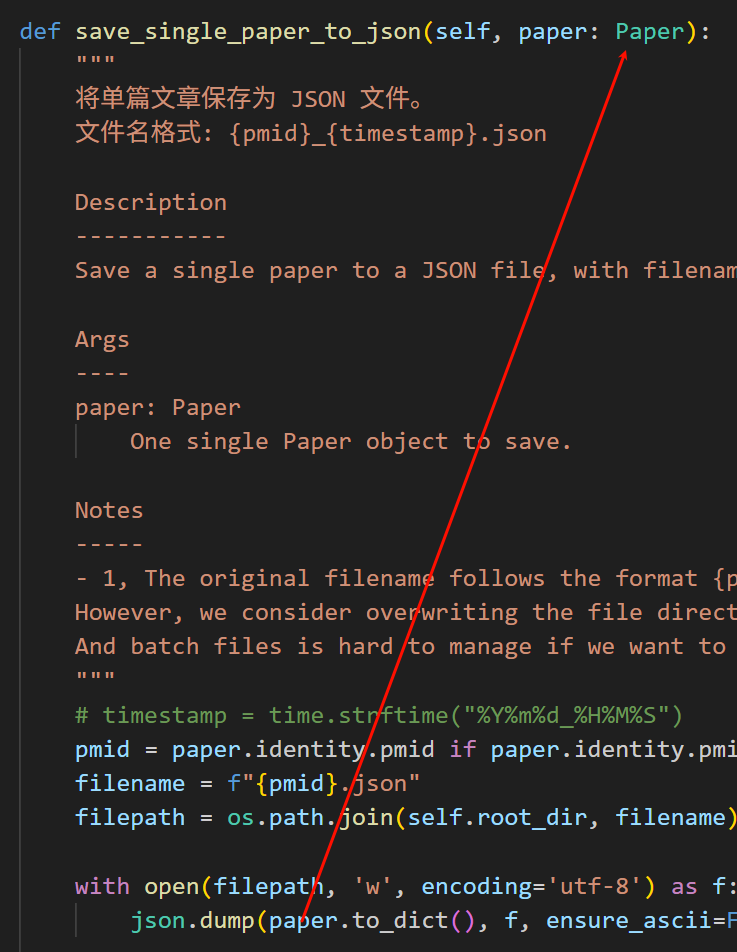
可以看到,我这里写的时候中途修改了数据类,本来是打算传入一个paper类,然后在paper类里实现一个todict的;
但是后面我又在paper类的基础上改了改,我将原本的paper类改为了paper_metadata类,
然后又重新在上一层抽象出了一个paper类;
所以这里原始的就没有改过来。
那这里就是简单的需求修改没有及时同步,我们再改过来试试:
还是报了同样的错,鉴于bug比较多,我只贴出最后相同的错误,先修最下面的要紧
python
-> Fetching external LinkOuts (Datasets, Full Text, etc.) for 12 PMIDs at [2025-12-19 19:01:33] ...
Fetching articles 1 to 12 failed: [DictionaryElement.__init__() missing 2 required positional arguments: 'attrs' and 'allowed_tags'] at [2025-12-19 19:01:35] ...
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 432, in fetch_from_query
self.save_single_paper_to_json(paper)
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~^^^^^^^
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 1320, in save_single_paper_to_json
json.dump(paper.to_dict(), f, ensure_ascii=False, sort_keys=True, indent=4)
~~~~~~~~~~~~~^^
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 157, in to_dict
return asdict(self)
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/dataclasses.py", line 1359, in asdict
return _asdict_inner(obj, dict_factory)
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/dataclasses.py", line 1370, in _asdict_inner
f.name: _asdict_inner(getattr(obj, f.name), dict)
~~~~~~~~~~~~~^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/dataclasses.py", line 1370, in _asdict_inner
f.name: _asdict_inner(getattr(obj, f.name), dict)
~~~~~~~~~~~~~^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/dataclasses.py", line 1380, in _asdict_inner
return [_asdict_inner(v, dict_factory) for v in obj]
~~~~~~~~~~~~~^^^^^^^^^^^^^^^^^
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/dataclasses.py", line 1419, in _asdict_inner
return obj_type((_asdict_inner(k, dict_factory),
_asdict_inner(v, dict_factory))
for k, v in obj.items())
TypeError: DictionaryElement.__init__() missing 2 required positional arguments: 'attrs' and 'allowed_tags'
Saving 0 papers to storage...
Done.可以发现,问题还是处在to_dict上面,这个时候我才注意到应该好好看看TypeError说了什么,
DictionaryElement,这个是BioPython里面字典的格式,
会不会是这个格式没办法用常规的todict来转化,其实我们可以用一个测试数据去检验一下todict函数行不行。
推测是BioPython的特殊类型数据问题:
Biopython 的 Entrez.read() 返回的不是标准的 dict 或 list,而是它自己定义的Bio.Entrez.Parser.DictionaryElement 或 ListElement。
这些类型在被 dataclasses.asdict 序列化时会出问题。
解决方法其实很简单:
- 要么就是不用asdict,看看有没有什么办法可以将BioPython返回的数据类转化为dict的专用函数
- 还是用asdict,但是在用之前先深度清洗一下,就是将其返回的数据转换为我们常用常见的数据格式
第一个解决方案,我去google以及biopython的ISSUE里看了下,没怎么找到回答。
所以暂时转向第2个解决方案,这就涉及到我解析Biopython的返回函数里面的逻辑了,
这里我写了3个函数用于解析Biopython中的返回值,
所以我需要在这3个函数中都check一遍具体提取的逻辑:
仔细看了下,前两个函数因为我需要深入数据分析,所以我是完全解析的,也就是字典、列表之类的元素我完全解析了出来,基本上都转换为了正常的数据格式。
只有第3个函数,也就是获取外部链接数据方面,因为键值对比较复杂,所以我就直接放弃了解析,
直接就将列表数据赋值给了我的返回结果中的一个变量,
结果在这个变量需要整体转换为dict的时候,牵扯到了这个没有被解析的Biopython的特殊原始数据格式,所以报错了。
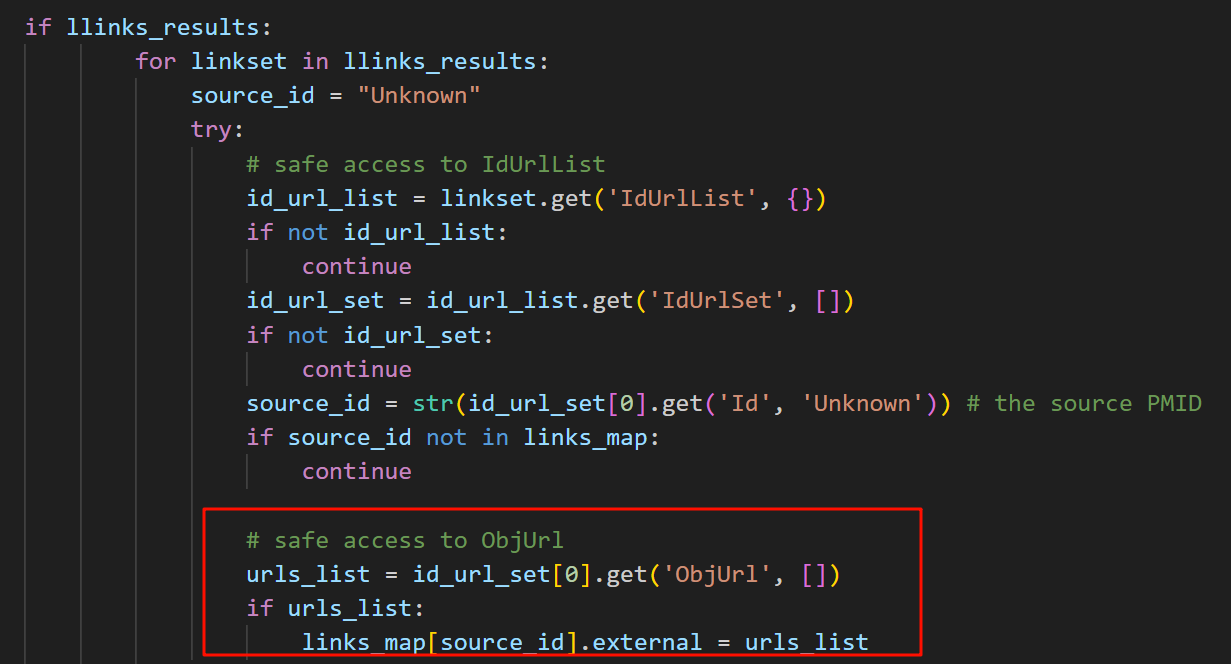
所以,我现在不得不回过头去再解析一下这个字典。
为了方便还原分析,我此处举一个相似的例子:
python
results[0]['IdUrlList']['IdUrlSet'][0]['ObjUrl']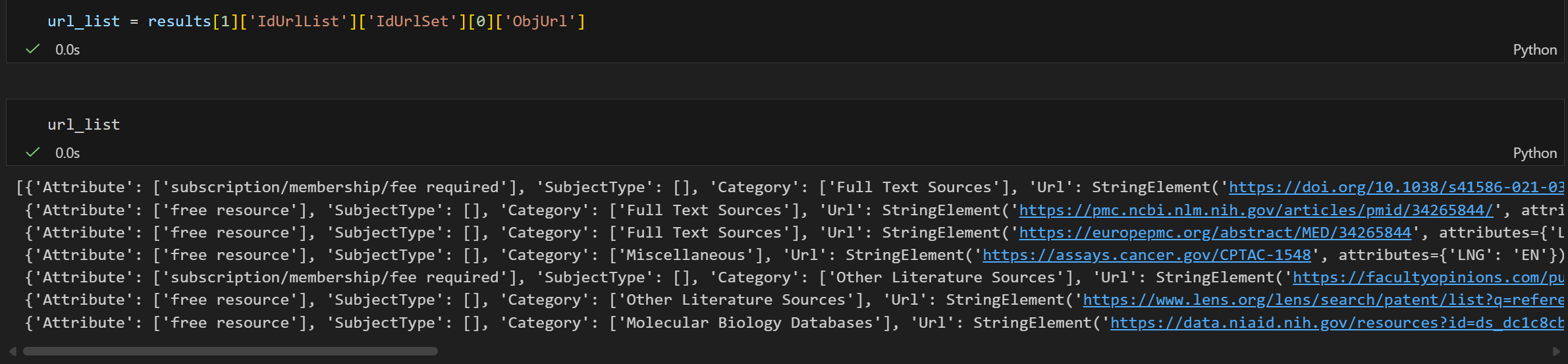
这个就相当于我们前面直接赋值给左边的那个biopython的数据类型,这里为了简洁说明,同样取名为url_list。
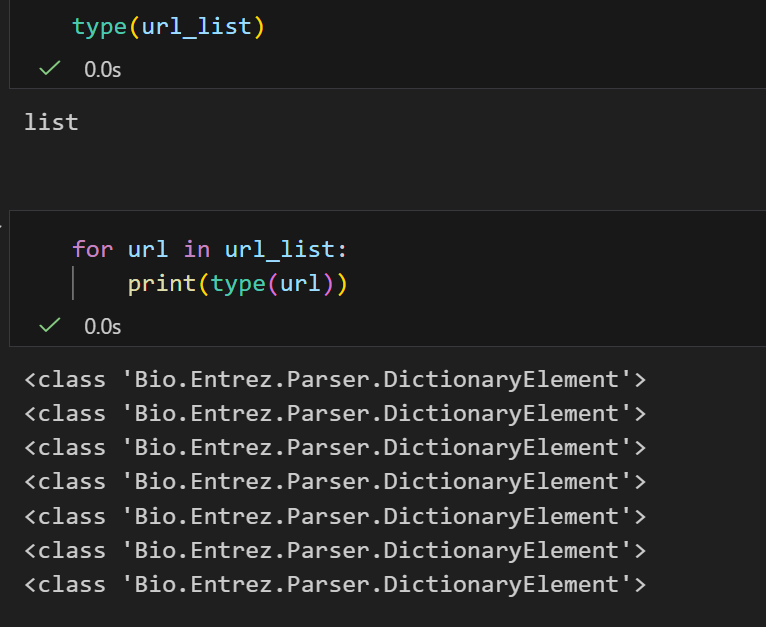
看了一下,确实是特殊的字典类型,对上了
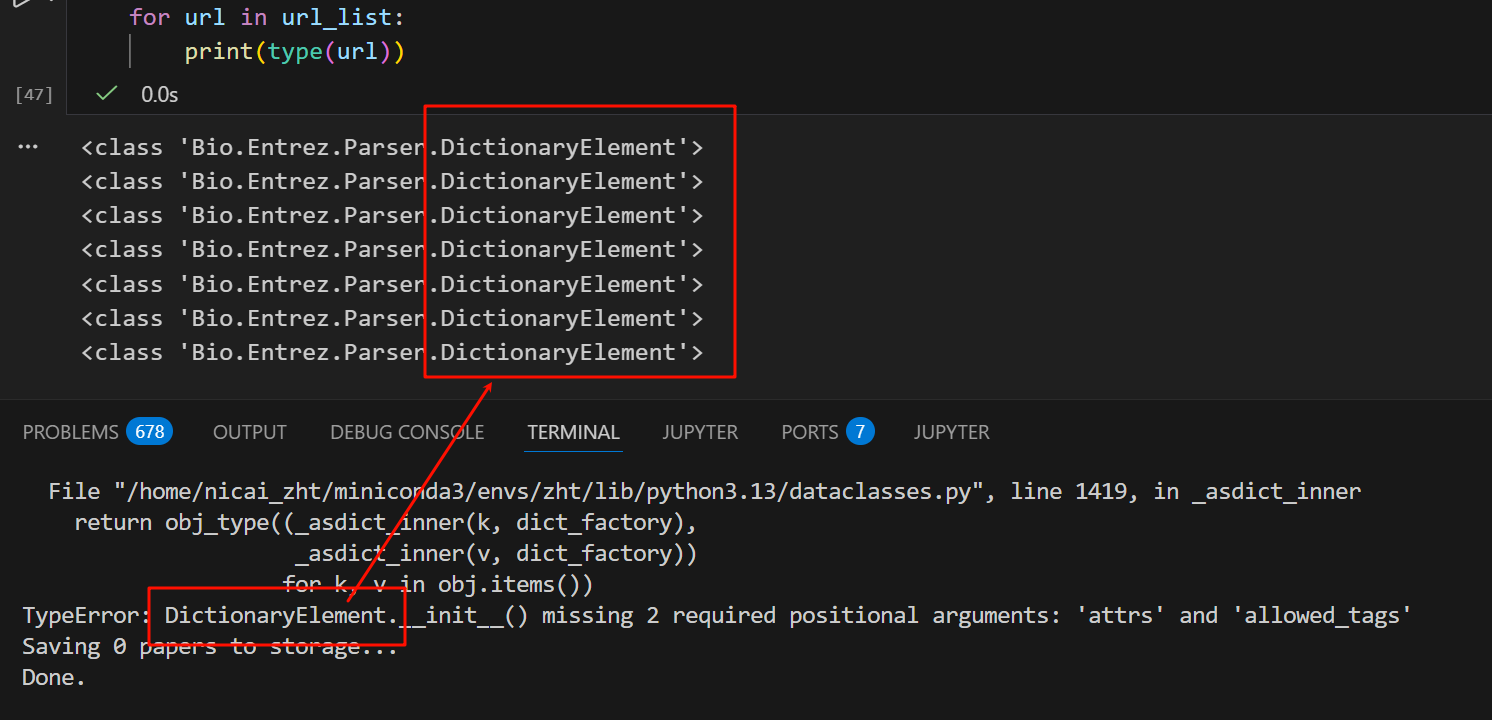
现在我们来简单测试一下,用这个url_list构建一个数据类dataclass
python
from dataclasses import asdict, dataclass
@dataclass
class Url:
Url: list
test1 = Url(url_list)
test1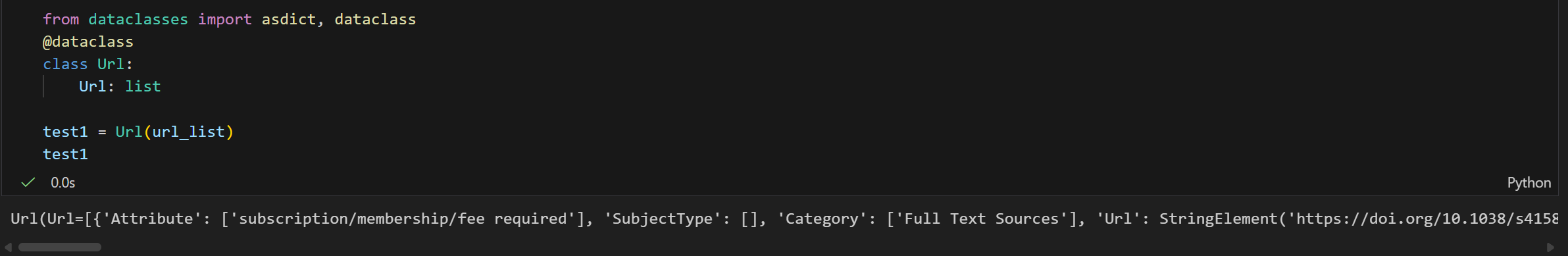
可以发现是一样的报错!
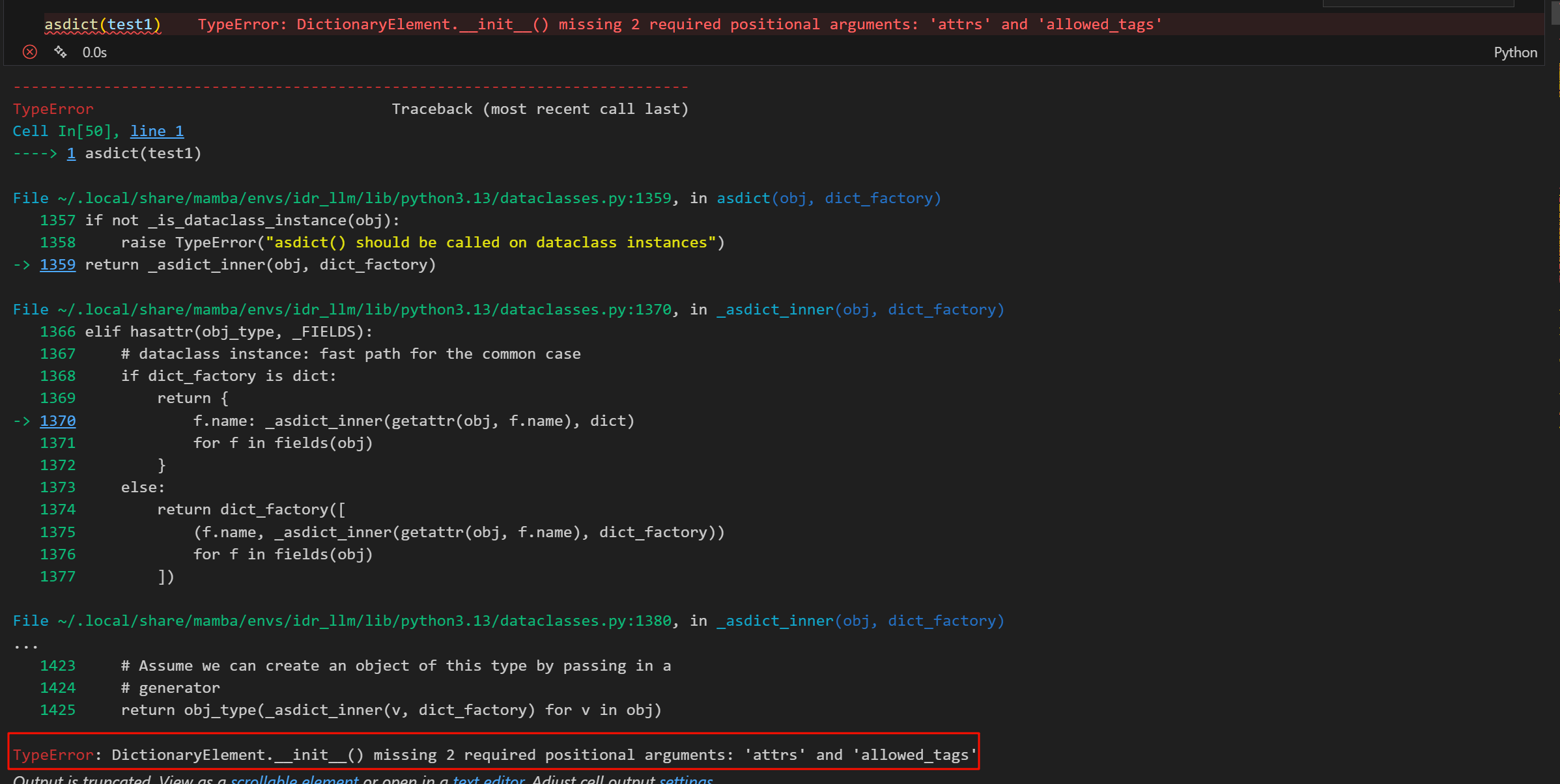
python
for url in url_list:
for key, value in url.items():
print(f"{key}: {value}")
print("-----"*10)简单看了一下:
发现其中的键值对还是相对固定的,
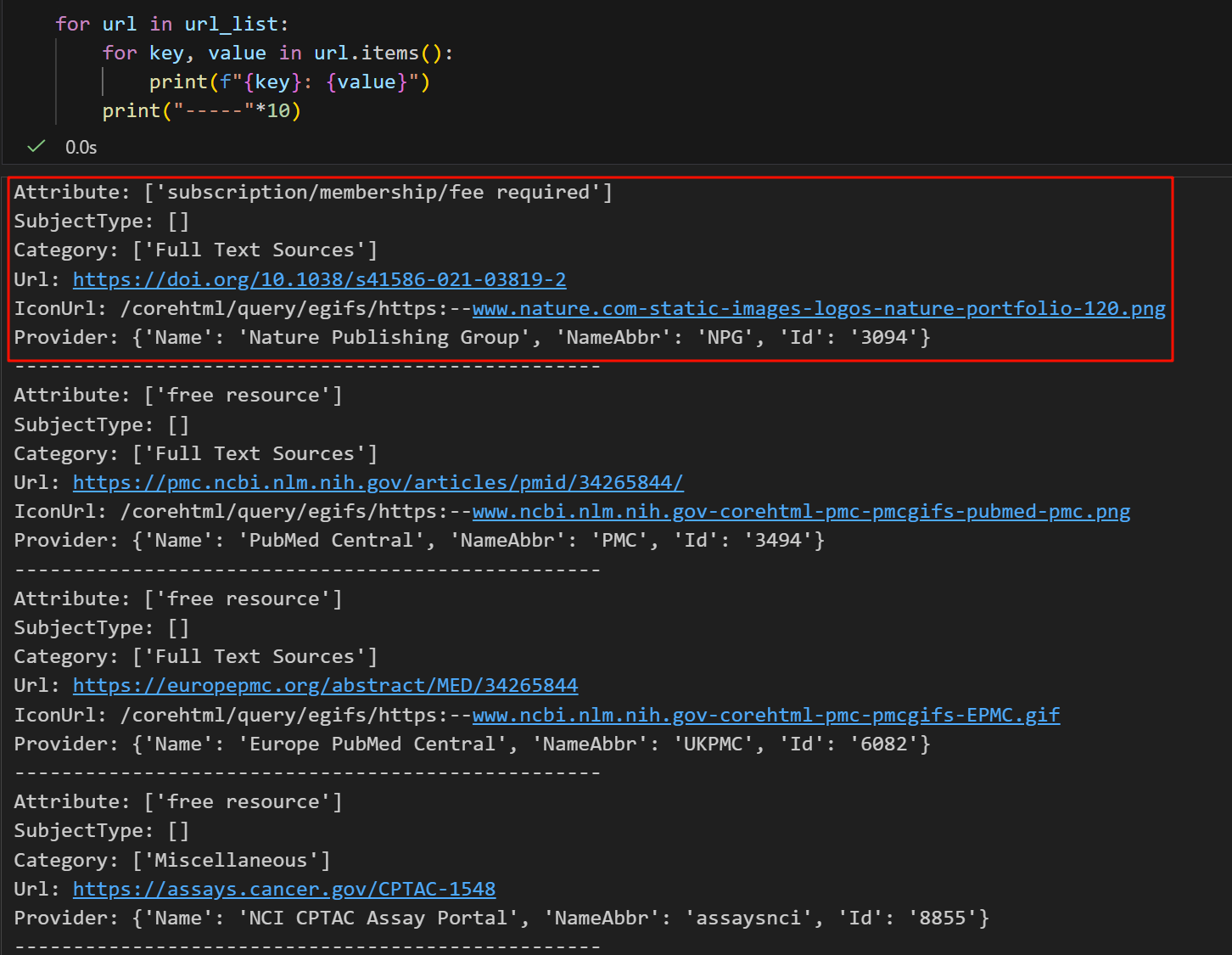
简单看了下,其中比较有用的数据大概是category、url、provider,Icon图表完全没用、attribute没用,但是subjecttype不知道是什么,其余有些有的有些没有的键值对可以做防御性编程。
总之,在正确解析数据之后再次运行:
先不看别的,至少这部分结果没问题了!
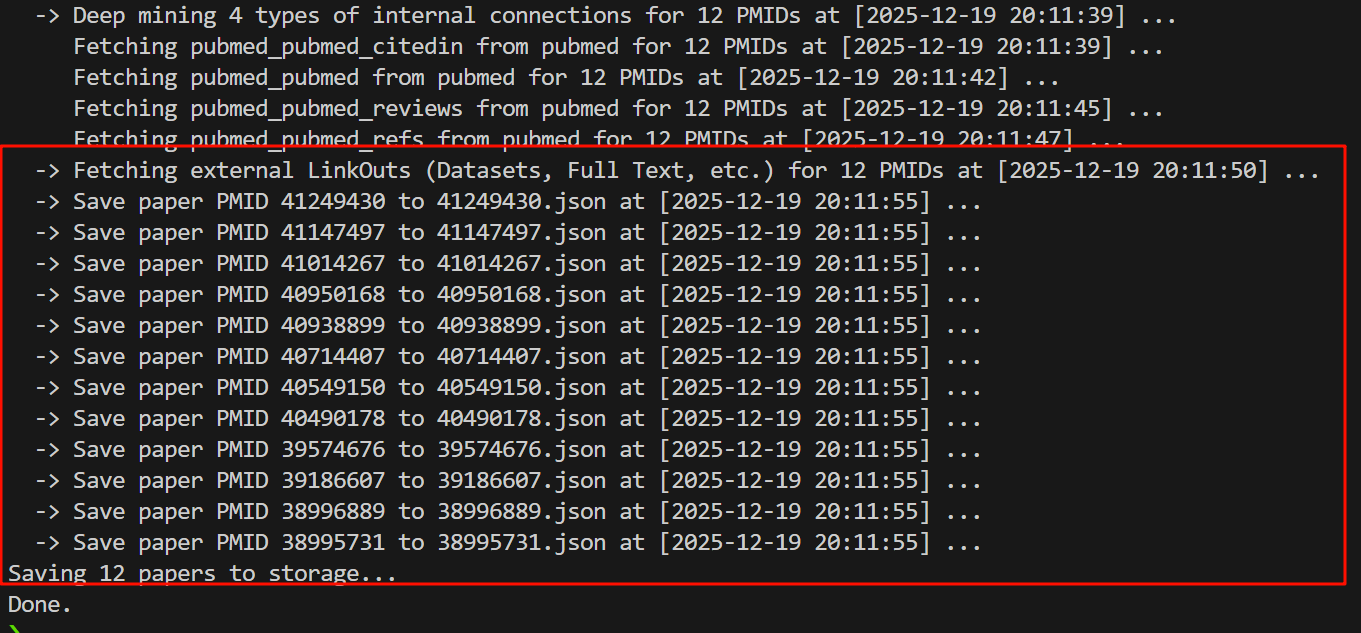
我们可以看一下返回的结果:
下面是其中一个结果:一个很复杂的Json格式文件
python
{
"content": {
"abstract": "MOTIVATION: Sidechain rotamer libraries of the common amino acids of a protein are useful for folded protein structure determination and for generating ensembles of intrinsically disordered proteins (IDPs). However, much of protein function is modulated beyond the translated sequence through the introduction of post-translational modifications (PTMs). RESULTS: In this work, we have provided a curated set of side chain rotamers for the most common PTMs derived from the RCSB PDB database, including phosphorylated, methylated, and acetylated sidechains. Our rotamer libraries improve upon existing methods such as SIDEpro, Rosetta, and AlphaFold3 in predicting the experimental structures for PTMs in folded proteins. In addition, we showcase our PTM libraries in full use by generating ensembles with the Monte Carlo Side Chain Entropy (MCSCE) for folded proteins, and combining MCSCE with the Local Disordered Region Sampling algorithms within IDPConformerGenerator for proteins with intrinsically disordered regions. AVAILABILITY AND IMPLEMENTATION: The codes for dihedral angle computations and library creation are available at https://github.com/THGLab/ptm_sc.git.",
"keywords": [],
"mesh_terms": [
"*Protein Processing, Post-Translational",
"*Proteins/chemistry/metabolism",
"*Databases, Protein",
"*Intrinsically Disordered Proteins/chemistry/metabolism",
"Algorithms",
"Protein Folding",
"Monte Carlo Method",
"Protein Conformation",
"Amino Acids/chemistry/metabolism",
"Software"
],
"pub_types": [
"Journal Article"
]
},
"contributors": {
"medline": {
"affiliations": [
"Kenneth S. Pitzer Center for Theoretical Chemistry, University of California, Berkeley, CA 94720, United States.",
"Department of Chemistry, University of California, Berkeley, CA 94720, United States.",
"Molecular Medicine Program, Hospital for Sick Children, Toronto, ON M5G 0A4, Canada.",
"Department of Biochemistry, University of Toronto, Toronto, ON M5S 1A8, Canada.",
"Molecular Medicine Program, Hospital for Sick Children, Toronto, ON M5G 0A4, Canada.",
"Department of Biochemistry, University of Toronto, Toronto, ON M5S 1A8, Canada.",
"Kenneth S. Pitzer Center for Theoretical Chemistry, University of California, Berkeley, CA 94720, United States.",
"Department of Chemistry, University of California, Berkeley, CA 94720, United States.",
"Department of Bioengineering, University of California, Berkeley, CA 94720, United States.",
"Department of Chemical and Biomolecular Engineering, University of California, Berkeley, CA 94720, United States."
],
"auids": [
"ORCID: 0000-0002-8357-8507",
"ORCID: 0000-0003-0025-8987"
],
"full_names": [
"Zhang, Oufan",
"Naik, Shubhankar A",
"Liu, Zi Hao",
"Forman-Kay, Julie",
"Head-Gordon, Teresa"
],
"short_names": [
"Zhang O",
"Naik SA",
"Liu ZH",
"Forman-Kay J",
"Head-Gordon T"
]
},
"xml": [
{
"affiliations": [
"Kenneth S. Pitzer Center for Theoretical Chemistry, University of California, Berkeley, CA 94720, United States."
],
"full_name": "Zhang, Oufan",
"identifiers": [],
"short_name": "Zhang O"
},
{
"affiliations": [
"Department of Chemistry, University of California, Berkeley, CA 94720, United States."
],
"full_name": "Naik, Shubhankar A",
"identifiers": [],
"short_name": "Naik SA"
},
{
"affiliations": [
"Molecular Medicine Program, Hospital for Sick Children, Toronto, ON M5G 0A4, Canada.",
"Department of Biochemistry, University of Toronto, Toronto, ON M5S 1A8, Canada."
],
"full_name": "Liu, Zi Hao",
"identifiers": [
"0000-0002-8357-8507"
],
"short_name": "Liu ZH"
},
{
"affiliations": [
"Molecular Medicine Program, Hospital for Sick Children, Toronto, ON M5G 0A4, Canada.",
"Department of Biochemistry, University of Toronto, Toronto, ON M5S 1A8, Canada."
],
"full_name": "Forman-Kay, Julie",
"identifiers": [],
"short_name": "Forman-Kay J"
},
{
"affiliations": [
"Kenneth S. Pitzer Center for Theoretical Chemistry, University of California, Berkeley, CA 94720, United States.",
"Department of Chemistry, University of California, Berkeley, CA 94720, United States.",
"Department of Bioengineering, University of California, Berkeley, CA 94720, United States.",
"Department of Chemical and Biomolecular Engineering, University of California, Berkeley, CA 94720, United States."
],
"full_name": "Head-Gordon, Teresa",
"identifiers": [
"0000-0003-0025-8987"
],
"short_name": "Head-Gordon T"
}
]
},
"identity": {
"doi": "10.1093/bioinformatics/btae444",
"pmid": "38995731",
"title": "A curated rotamer library for common post-translational modifications of proteins."
},
"links": {
"cites": [
"40799364",
"40789114",
"40093079",
"39279844",
"39062519"
],
"entrez": {},
"external": [
{
"attribute": "subscription/membership/fee required",
"category": "Full Text Sources",
"linkname": "",
"provider": "Ovid Technologies, Inc.",
"url": "http://ovidsp.ovid.com/ovidweb.cgi?T=JS&PAGE=linkout&SEARCH=38995731.ui"
},
{
"attribute": "free resource",
"category": "Full Text Sources",
"linkname": "",
"provider": "PubMed Central",
"url": "https://pmc.ncbi.nlm.nih.gov/articles/pmid/38995731/"
},
{
"attribute": "free resource",
"category": "Miscellaneous",
"linkname": "",
"provider": "NCI CPTAC Assay Portal",
"url": "https://assays.cancer.gov/CPTAC-1532"
},
{
"attribute": "free resource",
"category": "Full Text Sources",
"linkname": "Full text from University of California eScholarship",
"provider": "eScholarship, University of California - Access Free Full Text",
"url": "https://escholarship.org/uc/item/qt4fs236m4"
},
{
"attribute": "subscription/membership/fee required",
"category": "Full Text Sources",
"linkname": "",
"provider": "Silverchair Information Systems",
"url": "https://academic.oup.com/bioinformatics/article-lookup/doi/10.1093/bioinformatics/btae444"
}
],
"fetch_timestamp": "",
"full_text_pmc": [],
"refs": [
"38718835",
"38428397",
"38060268",
"37956700",
"37904608",
"37723259",
"37686234",
"37253017",
"37144719",
"36639584",
"36257022",
"36030416",
"35122328",
"34282049",
"34265844",
"33826699",
"33305318",
"32259206",
"31368302",
"30703322",
"30589846",
"30418626",
"28430426",
"27387657",
"26397464",
"25531225",
"25482539",
"24857970",
"24823488",
"24474795",
"22593013",
"22072531",
"21918110",
"21873640",
"21645855",
"21187238",
"18573080",
"16863191",
"15980494",
"10592235",
"9260279",
"9254694"
],
"review": [
"38995731",
"26851279",
"26851286",
"29982297",
"25307499",
"12163064",
"26922996",
"34390736",
"29100108",
"24532081",
"31694155",
"37219402",
"24889695",
"31441158",
"25502193"
],
"similar": [
"38995731",
"38764597",
"36030416",
"26851279",
"38060268",
"24574112",
"37686234",
"26851286",
"26851284",
"22072531",
"22108787",
"29626537",
"31613621",
"31851497",
"27111521",
"31375817",
"29982297",
"25307499",
"36327064",
"24354725",
"38796681",
"37439702",
"12163064",
"22544766",
"38781500",
"21280126",
"22279426",
"26922996",
"21645855",
"31362509",
"38461354",
"26742101",
"37546943",
"14978310",
"32357776",
"30204435",
"32230759",
"22373394",
"32525675",
"29544285",
"36350035",
"32696395",
"24857970",
"31411026",
"31161204",
"27018641",
"34788852",
"31425549",
"29187139",
"29573987",
"31756104",
"28934129",
"26138206",
"26307970",
"34390736",
"26195786",
"33582133",
"33810353",
"19775152",
"26994550",
"22962462",
"29100108",
"24532081",
"31694155",
"28150238",
"23754493",
"37219402",
"33945530",
"9325103",
"32696367",
"29595057",
"18076032",
"28970487",
"26618856",
"36257022",
"24889695",
"31441158",
"29990433",
"24174539",
"24888500",
"29734867",
"15728119",
"31479556",
"25361972",
"33230318",
"26418581",
"33481784",
"17391016",
"33237329",
"18831774",
"25502193",
"39250597",
"28365882",
"35883018",
"22095648",
"29490246",
"30184055",
"9080199",
"26374675",
"31415767",
"11469867"
]
},
"metadata": {
"entrez_date": "2024/07/12 18:42",
"fetched_at": "2025-12-19 20:11:22",
"source_format": "Medline"
},
"source": {
"journal_abbrev": [
"Bioinformatics"
],
"journal_title": [
"Bioinformatics (Oxford, England)"
],
"pub_date": "2024 Jul 1",
"pub_types": [
"Journal Article"
],
"pub_year": "2024"
}
}现在我们再回过头来查看前面的问题:
python
Error in batch Elink acheck for 12 PMIDs: [IncompleteRead(167 bytes read)] at [2025-12-19 21:44:47] ...
Traceback (most recent call last):
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/http/client.py", line 579, in _get_chunk_left
chunk_left = self._read_next_chunk_size()
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/http/client.py", line 546, in _read_next_chunk_size
return int(line, 16)
ValueError: invalid literal for int() with base 16: b''
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/http/client.py", line 597, in _read_chunked
while (chunk_left := self._get_chunk_left()) is not None:
~~~~~~~~~~~~~~~~~~~~^^
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/http/client.py", line 581, in _get_chunk_left
raise IncompleteRead(b'')
http.client.IncompleteRead: IncompleteRead(0 bytes read)
The above exception was the direct cause of the following exception:
Traceback (most recent call last):
File "/data2/pyPaperFlow/src/pyPaperFlow/fetcher.py", line 990, in fetch_linked_data_for_batch_pmid
acheck_results = Entrez.read(handle)
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/site-packages/Bio/Entrez/__init__.py", line 529, in read
record = handler.read(source)
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/site-packages/Bio/Entrez/Parser.py", line 405, in read
self.parser.ParseFile(stream)
~~~~~~~~~~~~~~~~~~~~~^^^^^^^^
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/http/client.py", line 473, in read
return self._read_chunked(amt)
~~~~~~~~~~~~~~~~~~^^^^^
File "/home/nicai_zht/miniconda3/envs/zht/lib/python3.13/http/client.py", line 609, in _read_chunked
raise IncompleteRead(b''.join(value)) from exc
http.client.IncompleteRead: IncompleteRead(167 bytes read)很典型的:http.client.IncompleteRead: IncompleteRead。
问题出在acheck_results = Entrez.read(handle),
我的程序向 NCBI 发送了 elink 请求(cmd="acheck"),NCBI 开始发送响应,但传输到一半(只读了 167 字节)连接就断开了,或者数据包不完整。这通常是因为网络波动、NCBI 服务器负载过高或者超时。
至于文件输出,是因为我用了try-except,虽然程序捕获了网络错误,以及我试着在出错的时候给这个变量acheck_results赋一个空列表值。
但是为了保险起见,我其实已经强制定义了几个必须挖掘的链接,
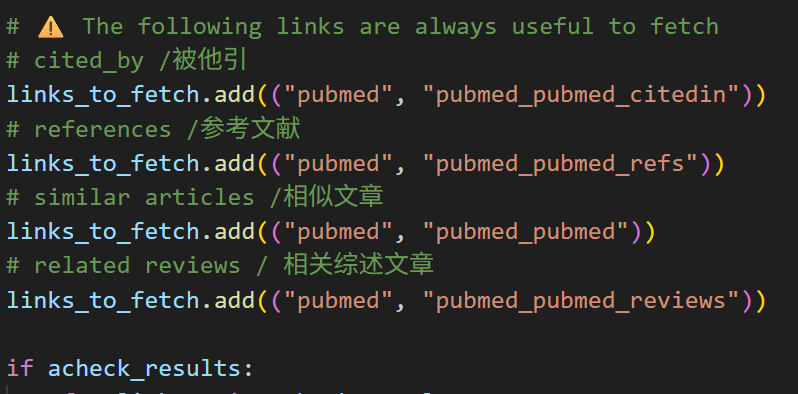
所以就算最下面的acheck_results失败了,但是我的链接列表也就是links_to_fetch里其实还要几个默认的固定链接要获取。只是这些文件里可能缺少了部分链接数据(因为 acheck 失败了)。
解决方法其实很简单,就是重试:
既然是网络波动,最好的办法是 "失败重试" (Retry)。
python
for attempt in range(max_retries):
# 再套try-except但是还是有问题,

只能说,这个可能是网络或者是NCBI的Entrez的问题,
导致我现在只能硬编码最重要的几个链接类型(引用、被引、相似文献),即使 acheck(发现阶段)彻底失败,我的程序依然会去抓取这些核心数据。acheck 失败仅仅意味着我可能漏掉了一些冷门的、非标准的链接类型,这对大多数分析来说无关紧要------------只能这么安慰我自己了。
既然是波动这种最玄学的问题,
那么就从请求频率、分析压力等玄学方面考虑,
改时间,5s也不行
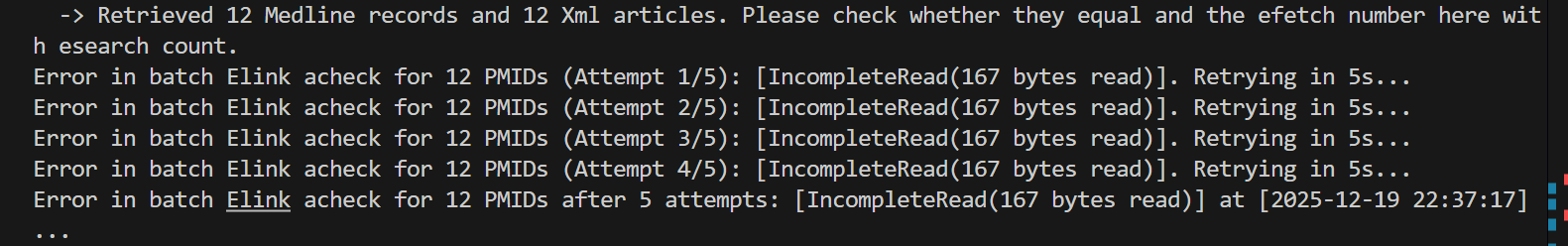
因为我是一批次50篇文献,有些是抓取元数据,这个比较快;
但是这个批次参数我也拿来用在获取外链,也就是我这里报错的波动上,报错的这个地方我用了elink的cmd=acheck模式,
可能是elink在cmd=acheck这个模式上计算太密集了,不好一次性处理我每批次50篇文献寻找所有外链存在性的请求。
解决方法可以是我再减少这个数目,就是每批次处理的数目,比如说在正式调用这个函数的时候,我可以10批次一个去检索之类,就是子切片。
现在我换成了10个一批:
前面10个依然挂了,但是后面这两个倒是成功了!

而且压力小的这两个拿到了更多的外链数据:整整多了4个,虽然看起来也没什么用
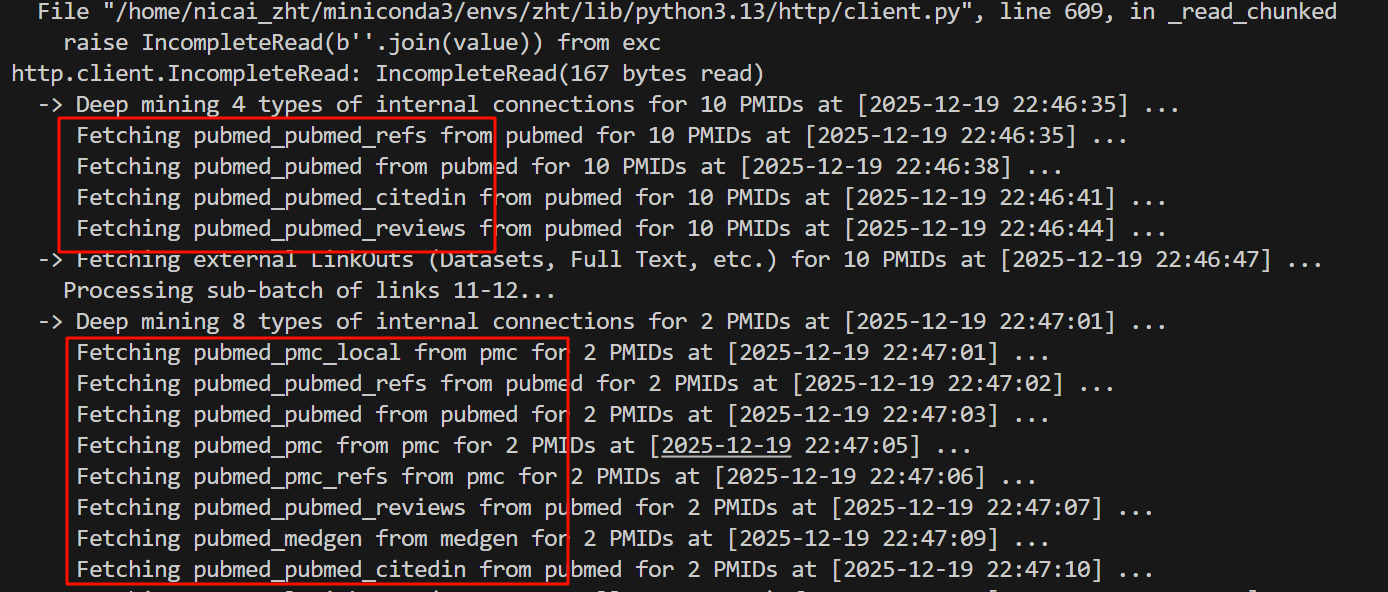
看来10个压力也大,干脆改成5个,
还是报错了:
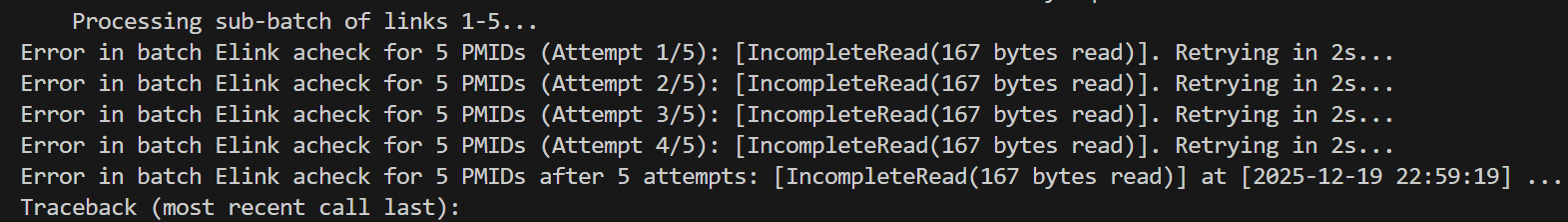
依旧只有后面2个成功:
但是如果再降低的话,那1~4,效率就很低了,而且我写的是batch处理函数,虽然也写了一个single脚本处理。
思索再三,想想看还是先别改 Batch Size 了,直接把 Traceback 注释掉。因为即使 acheck 失败,我依然拿到了最重要的引用关系和全文链接,这对后续分析已经足够了。不要为了 100% 的完美去和不稳定的 NCBI 服务器较劲。
后续总结:
- 把所有需要获取的链接都硬性编码
- 获取外链这一块就不要打印堆栈调用了,简短报错提示即可
- 得出结论:BioPython的Entrez模块的elink函数,在cmd="acheck"模式下,不能够处理批量pubmed的id,不然会因为网络波动等原因,极大概率挂掉,说明这种大型项目也不是神,维护也很困难,也有很多bug我们普通人都能够找出来