1、简介
文章地址:SegmentAnyBone: A Universal Model that Segments Any Bone at Any Location on MRI
这个分割一切骨头模型也是基于SAM的模型,官方在人体的195块骨头进行分割
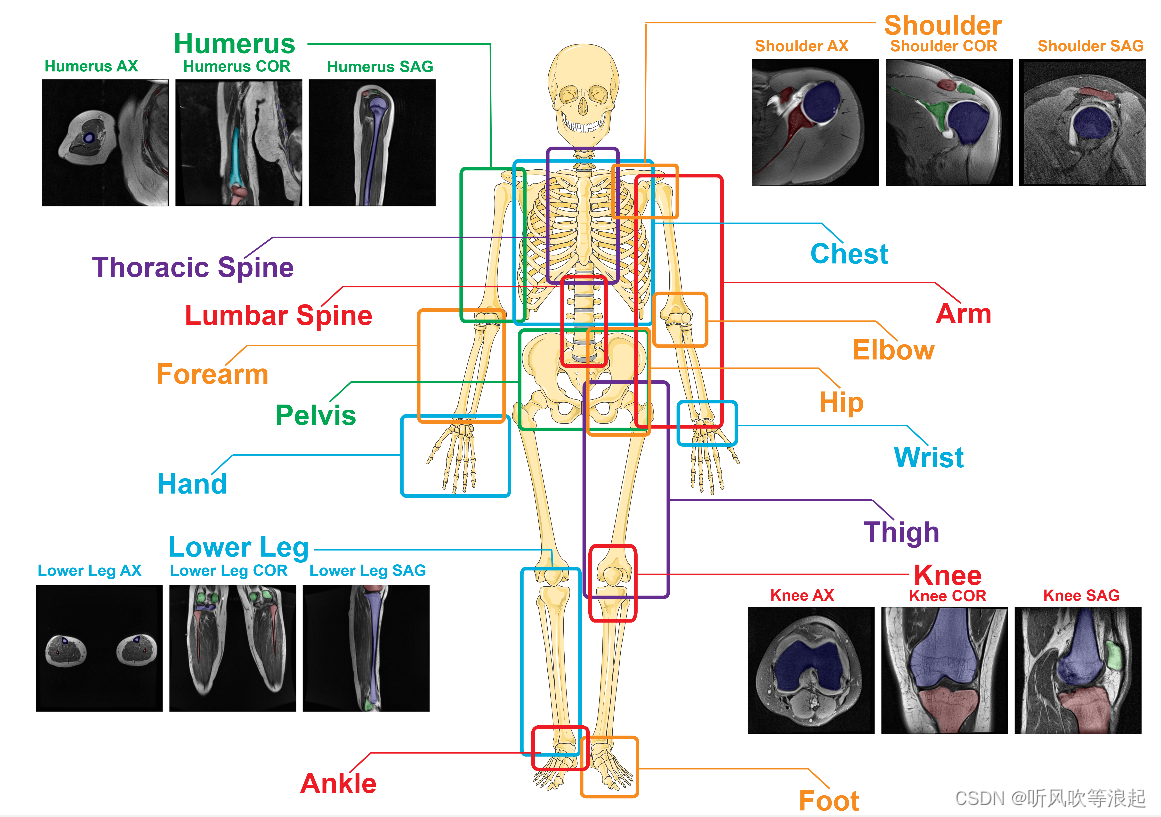
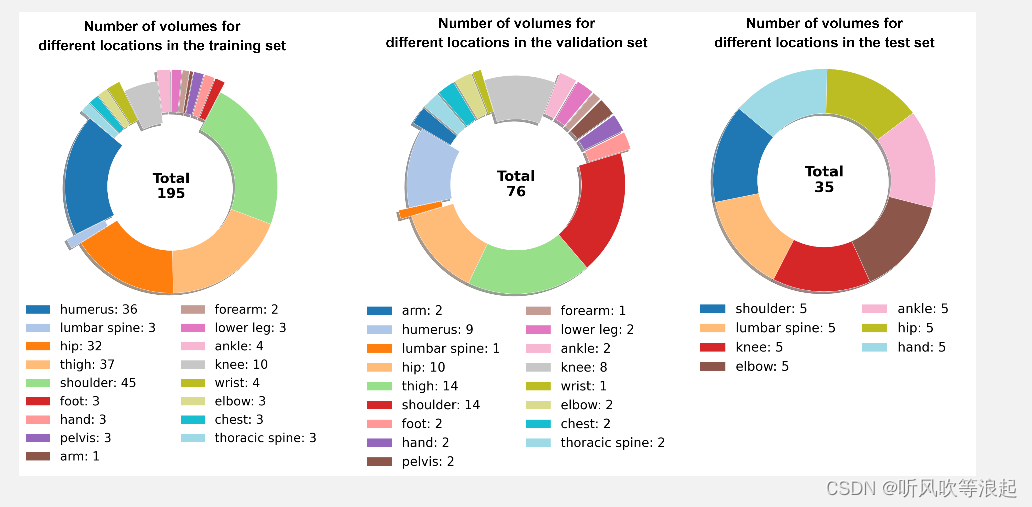
论文还没看,这里仅仅做复现
2、项目目录
项目下载的目录如下:
下载地址:SegmentAnyBone复现代码,包含2d、3d数据的推理和评估资源-CSDN文库
有些代码和目录是本人新加的,后面会介绍
这里不得不说,官方的代码和解释性很差,没有train的代码,对于推理,也只有一个ipynb的文件。所以这里仅仅介绍几种推理方法,推理的脚本是本人根据官方的提示重构的,仅供参考
3、准确工作
conda create -n segmentanybone python=3.10
激活后直接pip install -r requirements.txt 即可
预训练权重的下载:(总共三个)
MobileSAM/weights at master · ChaoningZhang/MobileSAM (github.com)

SegmentAnyBone Models - Google 云端硬盘

这里全部放在新建的checkpoint目录下

4、推理
官方给出的推理代码带有mask用于评估的,如果单纯的推理(没有gt的话),这里也简单做了实现
这里用py脚本实现,分为2D和3D的推理
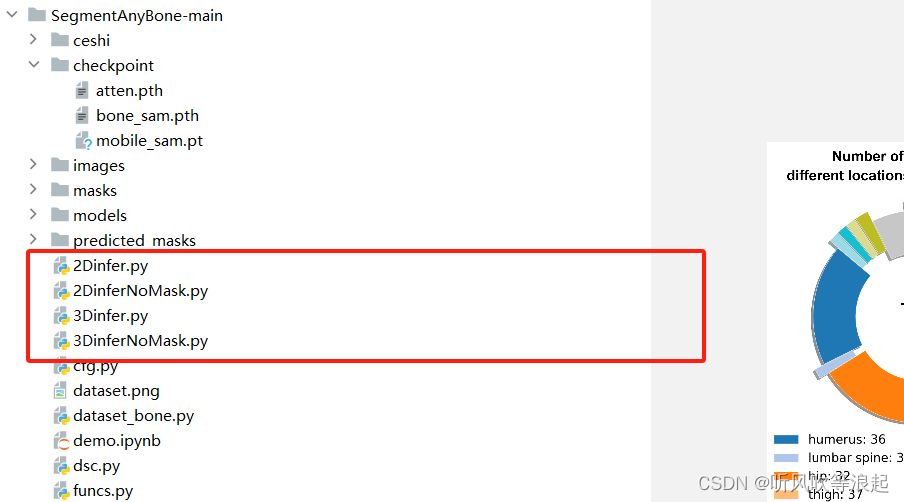
4.1 带有gt 的推理和评估
这里需要有数据和mask
4.1.1 数据摆放+转换(nii.gz to nrrd)
首先,数据集要放在images和masks目录下
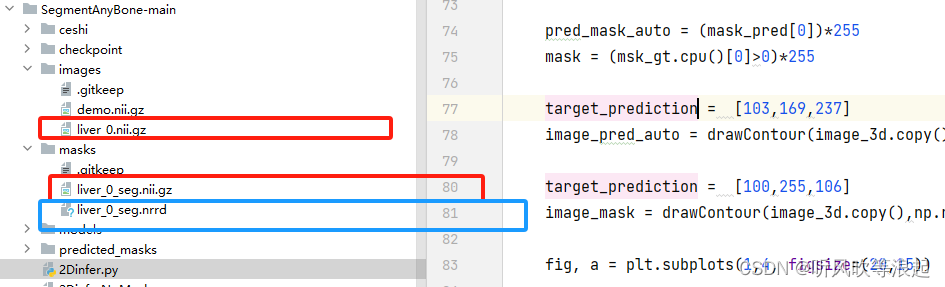
因为官方的代码用于MRI数据,所以这里的mask nii数据需要进行转换,可以参考下面的脚本:
python
import SimpleITK as sitk
def nii2nii(oripath,savepath):
data = sitk.ReadImage(oripath)
img = sitk.GetArrayFromImage(data)
out = sitk.GetImageFromArray(img)
sitk.WriteImage(out,savepath)
niitonrrd = nii2nii('./masks/liver_0_seg.nii.gz','liver_0_seg.nrrd')把生成的nrrd文件放在下面masks目录下即可
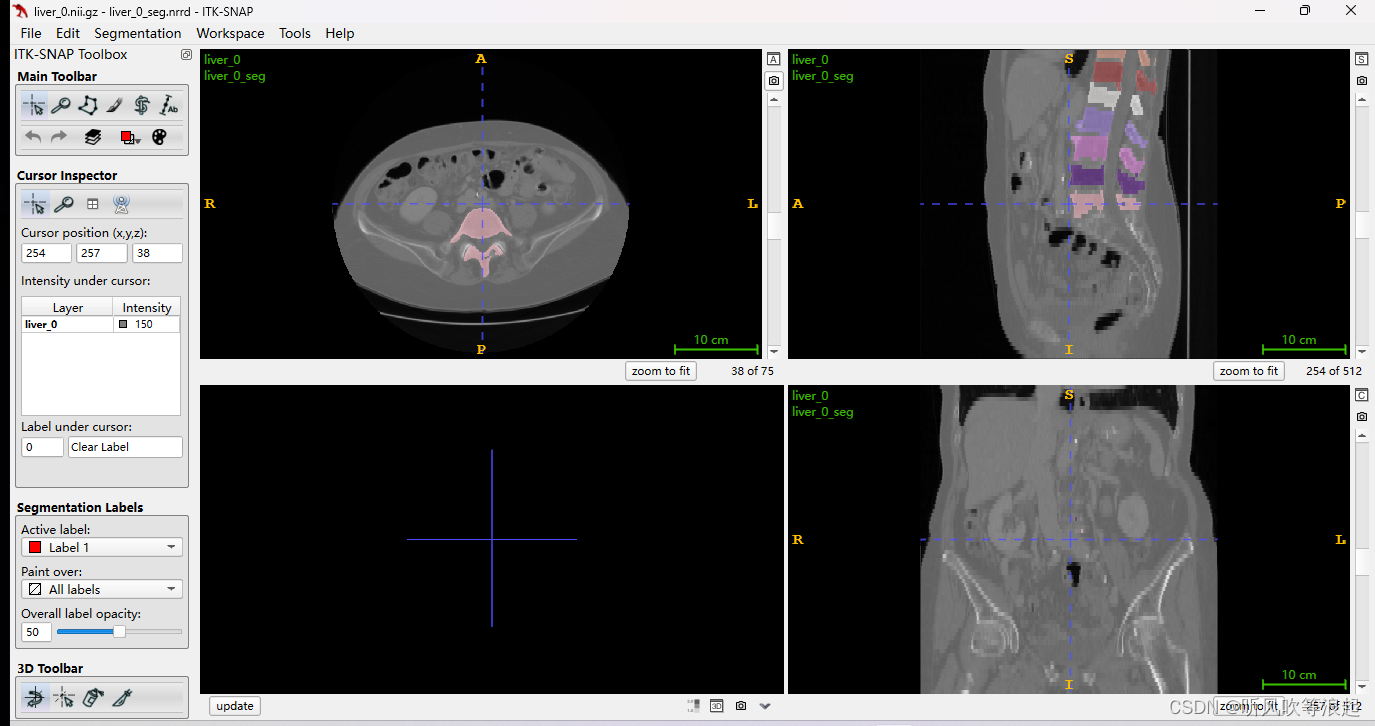
4.1.2 数据可视化
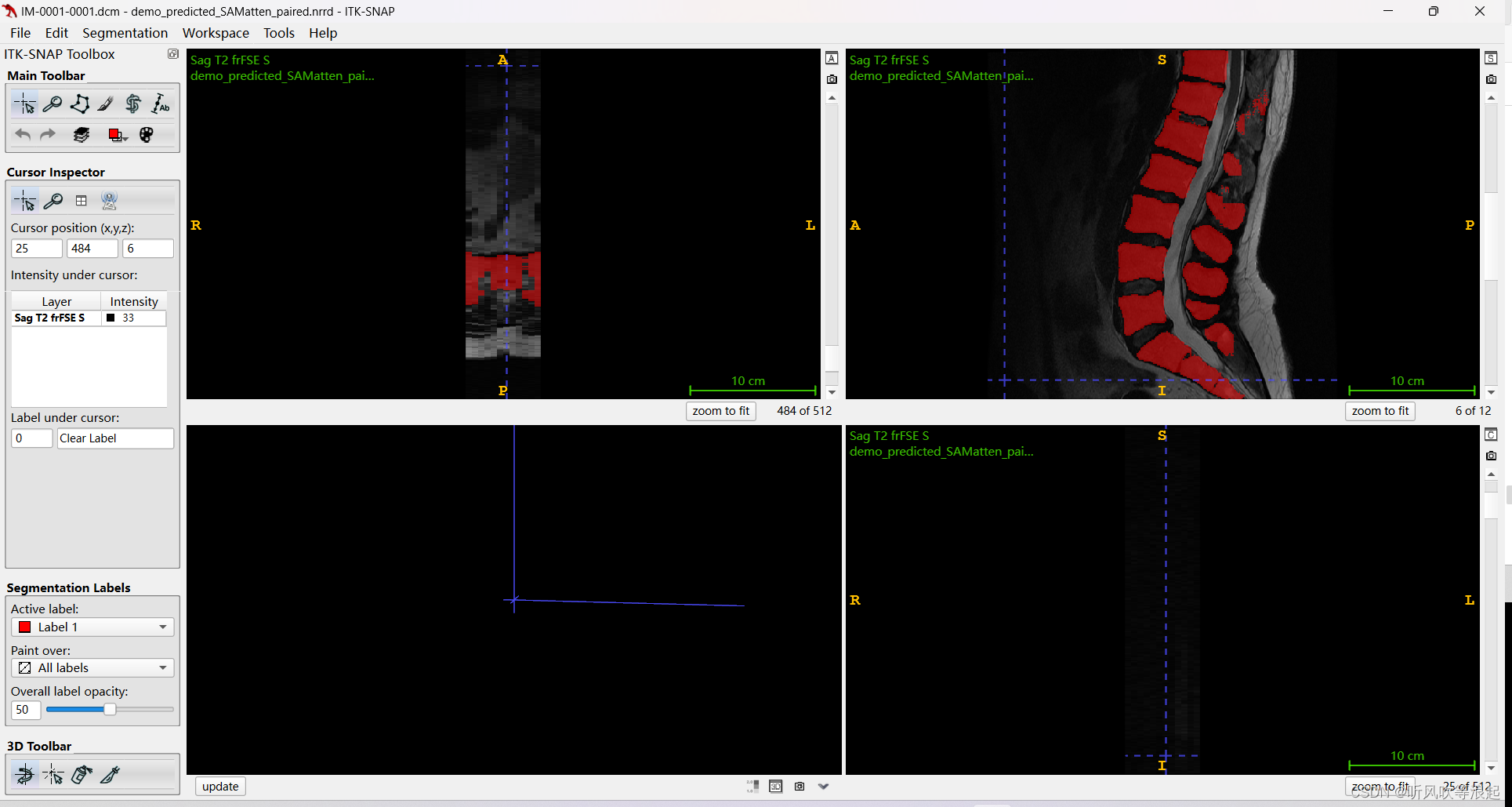
这里用ITK-SNAP可以可视化,mask用我们转换好的nrrd也是一样的效果
4.1.3 2D推理
代码如下:
python
from models.sam import SamPredictor, sam_model_registry
from models.sam.modeling.prompt_encoder import attention_fusion
import numpy as np
import os
import torch
import torchvision
import matplotlib.pyplot as plt
from torchvision import transforms
from PIL import Image
import matplotlib.pyplot as plt
from pathlib import Path
from dsc import dice_coeff
import torchio as tio
import nrrd
import PIL
import cfg
from funcs import *
from predict_funs import *
args = cfg.parse_args()
from monai.networks.nets import VNet
args.if_mask_decoder_adapter=True
args.if_encoder_adapter = True
args.decoder_adapt_depth = 2
def evaluateSlicePrediction(mask_pred, mask_name, slice_id):
voxels, header = nrrd.read(os.path.join(gt_msk_folder,mask_name))
mask_gt = voxels
msk = Image.fromarray(mask_gt[:,:,slice_id].astype(np.uint8), 'L')
msk = transforms.Resize((256,256))(msk)
msk_gt = (transforms.ToTensor()(msk)>0).float()
dsc_gt = dice_coeff(mask_pred.cpu(), msk_gt).item()
print("dsc_gt:", dsc_gt)
return msk_gt, dsc_gt
def predictSlice(image_name, lower_percentile, upper_percentile, slice_id, attention_enabled):
image1_vol = tio.ScalarImage(os.path.join(img_folder, image_name))
print('vol shape: %s vol spacing %s' %(image1_vol.shape,image1_vol.spacing))
image_tensor = image1_vol.data
lower_bound = torch_percentile(image_tensor, lower_percentile)
upper_bound = torch_percentile(image_tensor, upper_percentile)
# Clip the data
image_tensor = torch.clamp(image_tensor, lower_bound, upper_bound)
# Normalize the data to [0, 1]
image_tensor = (image_tensor - lower_bound) / (upper_bound - lower_bound)
image1_vol.set_data(image_tensor)
atten_map= pred_attention(image1_vol,vnet,slice_id,device)
atten_map = torch.unsqueeze(torch.tensor(atten_map),0).float().to(device)
print(atten_map.device)
if attention_enabled:
ori_img,pred_1,voxel_spacing1,Pil_img1,slice_id1 = evaluate_1_volume_withattention(image1_vol,sam_fine_tune,device,slice_id=slice_id,atten_map=atten_map)
else:
ori_img,pred_1,voxel_spacing1,Pil_img1,slice_id1 = evaluate_1_volume_withattention(image1_vol,sam_fine_tune,device,slice_id=slice_id)
mask_pred = ((pred_1>0)==cls).float().cpu()
return ori_img, mask_pred, atten_map
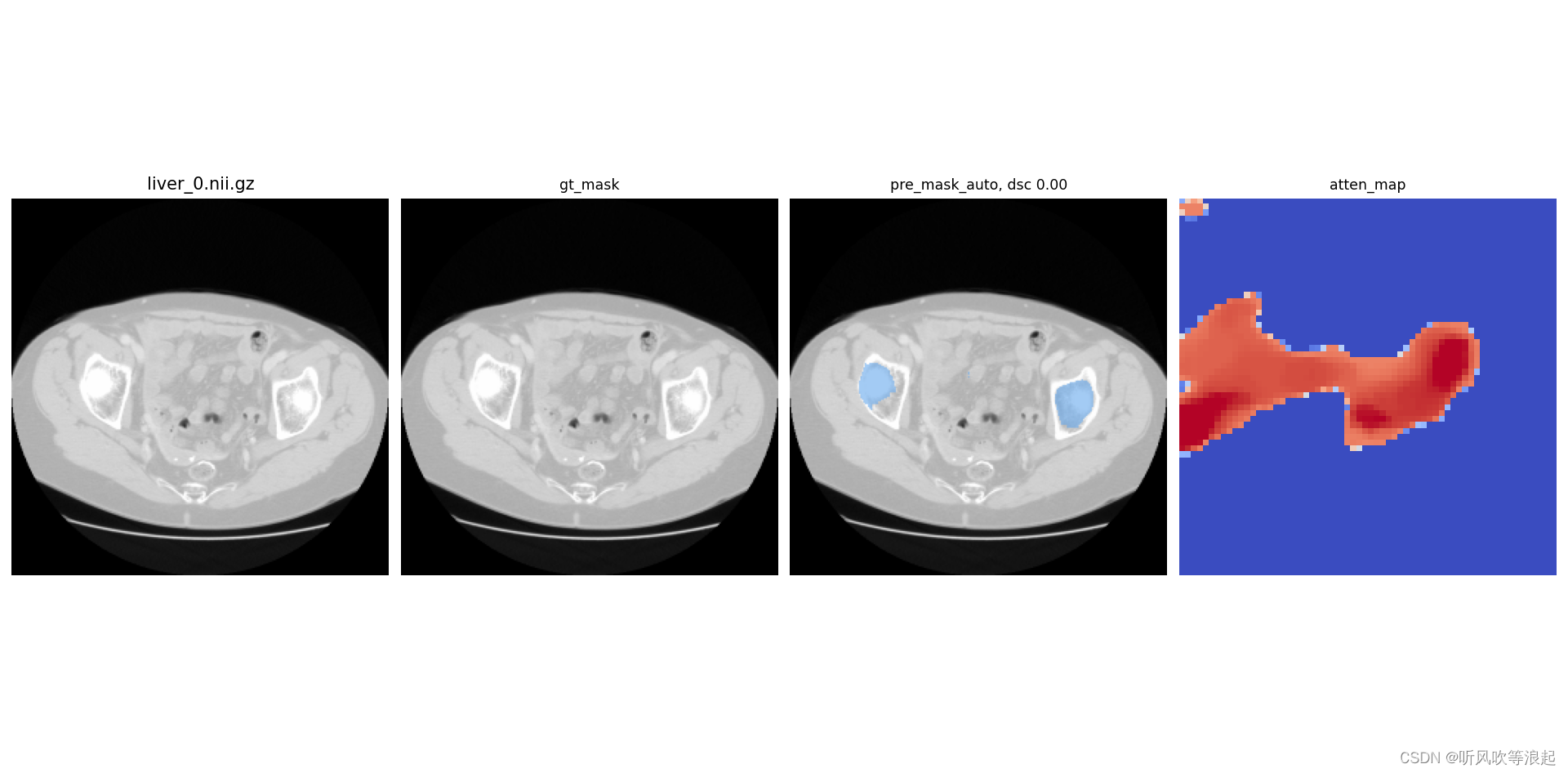
def visualizeSlicePrediction(ori_img, image_name, atten_map, msk_gt, mask_pred, dsc_gt):
image = np.rot90(torchvision.transforms.Resize((args.out_size,args.out_size))(ori_img)[0])
image_3d = np.repeat(np.array(image*255,dtype=np.uint8).copy()[:, :, np.newaxis], 3, axis=2)
pred_mask_auto = (mask_pred[0])*255
mask = (msk_gt.cpu()[0]>0)*255
target_prediction = [103,169,237]
image_pred_auto = drawContour(image_3d.copy(), np.rot90(pred_mask_auto),target_prediction,size=-1,a=0.6)
target_prediction = [100,255,106]
image_mask = drawContour(image_3d.copy(),np.rot90(mask),target_prediction,size=-1,a=0.6)
fig, a = plt.subplots(1,4, figsize=(20,15))
a[0].imshow(image,cmap='gray',vmin=0, vmax=1)
a[0].set_title(image_name)
a[0].axis(False)
a[1].imshow(image_mask,cmap='gray',vmin=0, vmax=255)
a[1].set_title('gt_mask',fontsize=10)
a[1].axis(False)
a[2].imshow(image_pred_auto,cmap='gray',vmin=0, vmax=255)
a[2].set_title('pre_mask_auto, dsc %.2f'%(dsc_gt),fontsize=10)
a[2].axis(False)
a[3].imshow(np.rot90(atten_map.cpu()[0]),vmin=0, vmax=1,cmap='coolwarm')
a[3].set_title('atten_map',fontsize=10)
a[3].axis(False)
plt.tight_layout()
plt.show()
device = 'cuda' if torch.cuda.is_available() else 'cpu'
print(device)
checkpoint_directory = './checkpoint' # path to your checkpoint
img_folder = os.path.join('images')
gt_msk_folder = os.path.join('masks')
predicted_msk_folder = os.path.join('predicted_masks')
cls = 1
sam_fine_tune = sam_model_registry["vit_t"](args,checkpoint=os.path.join(checkpoint_directory,'mobile_sam.pt'),num_classes=2)
sam_fine_tune.attention_fusion = attention_fusion()
sam_fine_tune.load_state_dict(torch.load(os.path.join(checkpoint_directory,'bone_sam.pth'),map_location=torch.device(device)), strict = True)
sam_fine_tune = sam_fine_tune.to(device).eval()
vnet = VNet().to(device)
model_directory = './checkpoint'
vnet.load_state_dict(torch.load(os.path.join(model_directory,'atten.pth'),map_location=torch.device(device)))
ori_img, predictedSliceMask, atten_map = predictSlice(
image_name = 'liver_0.nii.gz',
lower_percentile = 1,
upper_percentile = 99,
slice_id = 18, # slice number
attention_enabled = True, # if you want to use the depth attention
)
msk_gt, dsc_gt = evaluateSlicePrediction(
mask_pred = predictedSliceMask,
mask_name = 'liver_0_seg.nrrd',
slice_id = 18
)
visualizeSlicePrediction(
ori_img=ori_img,
image_name='liver_0.nii.gz',
atten_map=atten_map,
msk_gt=msk_gt,
mask_pred=predictedSliceMask,
dsc_gt=dsc_gt
)推理会展示可视化的效果图,并且计算推理和gt的dice值,这里的gt_mask是mask+image的掩模图

4.1.4 3D推理
代码如下:
python
from models.sam import SamPredictor, sam_model_registry
from models.sam.modeling.prompt_encoder import attention_fusion
import numpy as np
import os
import torch
import torchvision
import matplotlib.pyplot as plt
from torchvision import transforms
from PIL import Image
import matplotlib.pyplot as plt
from pathlib import Path
from dsc import dice_coeff
import torchio as tio
import nrrd
import PIL
import cfg
from funcs import *
from predict_funs import *
args = cfg.parse_args()
from monai.networks.nets import VNet
args.if_mask_decoder_adapter=True
args.if_encoder_adapter = True
args.decoder_adapt_depth = 2
def predictVolume(image_name, lower_percentile, upper_percentile):
dsc_gt = 0
image1_vol = tio.ScalarImage(os.path.join(img_folder,image_name))
print('vol shape: %s vol spacing %s' %(image1_vol.shape,image1_vol.spacing))
# Define the percentiles
image_tensor = image1_vol.data
lower_bound = torch_percentile(image_tensor, lower_percentile)
upper_bound = torch_percentile(image_tensor, upper_percentile)
# Clip the data
image_tensor = torch.clamp(image_tensor, lower_bound, upper_bound)
# Normalize the data to [0, 1]
image_tensor = (image_tensor - lower_bound) / (upper_bound - lower_bound)
image1_vol.set_data(image_tensor)
mask_vol_numpy = np.zeros(image1_vol.shape)
id_list = list(range(image1_vol.shape[3]))
for id in id_list:
atten_map = pred_attention(image1_vol,vnet,id,device)
atten_map = torch.unsqueeze(torch.tensor(atten_map),0).float().to(device)
ori_img,pred_1,voxel_spacing1,Pil_img1,slice_id1 = evaluate_1_volume_withattention(image1_vol,sam_fine_tune,device,slice_id=id,atten_map=atten_map)
img1_size = Pil_img1.size
mask_pred = ((pred_1>0)==cls).float().cpu()
pil_mask1 = Image.fromarray(np.array(mask_pred[0],dtype=np.uint8),'L').resize(img1_size,resample= PIL.Image.NEAREST)
mask_vol_numpy[0,:,:,id] = np.asarray(pil_mask1)
mask_vol = tio.LabelMap(tensor=torch.tensor(mask_vol_numpy,dtype=torch.int), affine=image1_vol.affine)
mask_save_folder = os.path.join(predicted_msk_folder,'/'.join(image_name.split('/')[:-1]))
Path(mask_save_folder).mkdir(parents=True, exist_ok = True)
mask_vol.save(os.path.join(mask_save_folder,image_name.split('/')[-1].replace('.nii.gz','_predicted_SAMatten_paired.nrrd')))
return mask_vol
def predictAndEvaluateVolume(image_name, mask_name, lower_percentile, upper_percentile):
dsc_gt = 0
image1_vol = tio.ScalarImage(os.path.join(img_folder,image_name))
print('vol shape: %s vol spacing %s' %(image1_vol.shape,image1_vol.spacing))
# Define the percentiles
image_tensor = image1_vol.data
lower_bound = torch_percentile(image_tensor, lower_percentile)
upper_bound = torch_percentile(image_tensor, upper_percentile)
# Clip the data
image_tensor = torch.clamp(image_tensor, lower_bound, upper_bound)
# Normalize the data to [0, 1]
image_tensor = (image_tensor - lower_bound) / (upper_bound - lower_bound)
image1_vol.set_data(image_tensor)
voxels, header = nrrd.read(os.path.join(gt_msk_folder,mask_name))
mask_gt = voxels
mask_vol_numpy = np.zeros(image1_vol.shape)
id_list = list(range(image1_vol.shape[3]))
for id in id_list:
atten_map = pred_attention(image1_vol,vnet,id,device)
atten_map = torch.unsqueeze(torch.tensor(atten_map),0).float().to(device)
ori_img,pred_1,voxel_spacing1,Pil_img1,slice_id1 = evaluate_1_volume_withattention(image1_vol,sam_fine_tune,device,slice_id=id,atten_map=atten_map)
img1_size = Pil_img1.size
mask_pred = ((pred_1>0)==cls).float().cpu()
msk = Image.fromarray(mask_gt[:,:,id].astype(np.uint8), 'L')
msk = transforms.Resize((256,256))(msk)
msk_gt = (transforms.ToTensor()(msk)>0).float().cpu()
dsc_gt += dice_coeff(mask_pred.cpu(),msk_gt).item()
pil_mask1 = Image.fromarray(np.array(mask_pred[0],dtype=np.uint8),'L').resize(img1_size,resample= PIL.Image.NEAREST)
mask_vol_numpy[0,:,:,id] = np.asarray(pil_mask1)
mask_vol = tio.LabelMap(tensor=torch.tensor(mask_vol_numpy,dtype=torch.int), affine=image1_vol.affine)
mask_save_folder = os.path.join(predicted_msk_folder,'/'.join(image_name.split('/')[:-1]))
Path(mask_save_folder).mkdir(parents=True,exist_ok = True)
mask_vol.save(os.path.join(mask_save_folder,image_name.split('/')[-1].replace('.nii.gz','_predicted_SAMatten_paired.nrrd')))
dsc_gt /= len(id_list)
gt_vol = tio.LabelMap(tensor=torch.unsqueeze(torch.Tensor(mask_gt>0),0), affine=image1_vol.affine)
dsc_vol = dice_coeff(mask_vol.data.float().cpu(),gt_vol.data).item()
print('volume %s: slice_wise_dsc %.2f; vol_wise_dsc %.2f'%(image_name,dsc_gt,dsc_vol))
device = 'cuda' if torch.cuda.is_available() else 'cpu'
print(device)
checkpoint_directory = './checkpoint' # path to your checkpoint
img_folder = os.path.join('images')
gt_msk_folder = os.path.join('masks')
predicted_msk_folder = os.path.join('predicted_masks')
cls = 1
sam_fine_tune = sam_model_registry["vit_t"](args,checkpoint=os.path.join(checkpoint_directory,'mobile_sam.pt'),num_classes=2)
sam_fine_tune.attention_fusion = attention_fusion()
sam_fine_tune.load_state_dict(torch.load(os.path.join(checkpoint_directory,'bone_sam.pth'),map_location=torch.device(device)), strict = True)
sam_fine_tune = sam_fine_tune.to(device).eval()
vnet = VNet().to(device)
model_directory = './checkpoint'
vnet.load_state_dict(torch.load(os.path.join(model_directory,'atten.pth'),map_location=torch.device(device)))
mask = predictVolume(
image_name = 'liver_0.nii.gz',
lower_percentile = 1,
upper_percentile = 99
)
predictAndEvaluateVolume(
image_name = 'liver_0.nii.gz',
mask_name = 'liver_0_seg.nrrd',
lower_percentile = 1,
upper_percentile = 99
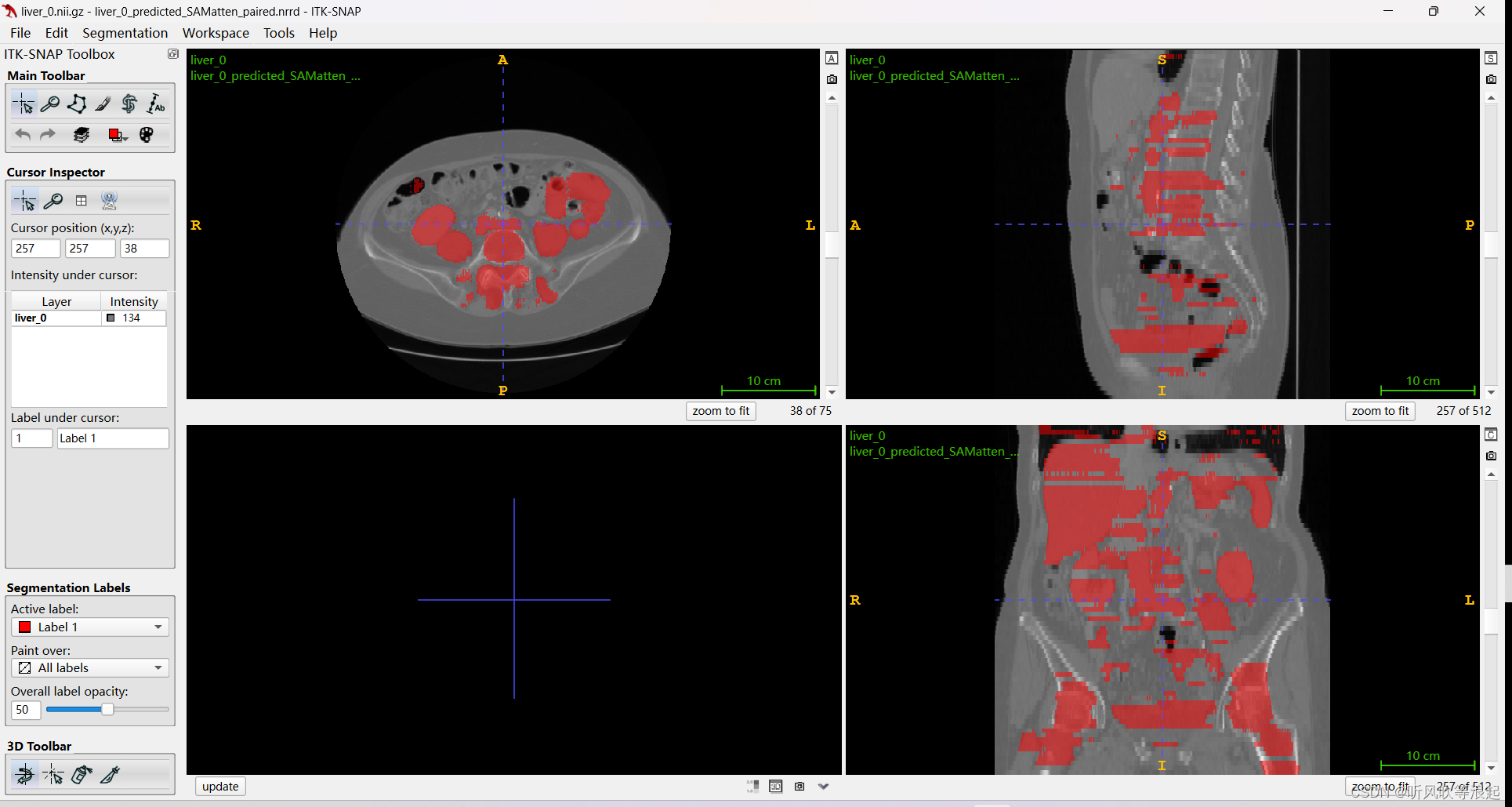
)推理完会生成结果的3d数据
效果很差,可能数据问题
4.2 没有mask的推理
这里其实就是nii的数据,没有对应的gt(nii.gz)而已
4.2.1 数据摆放+转换(dcm to nii.gz)
因为官方的代码是在MRI数据进行训练的,这里使用的也是骨头的MRI数据,不过是dcm的,需要将dcm的序列转为nii的3D数据

这个dcm数据也可以用ITK打开的
这种数据当然可以直接保存,然后转为nii格式就行了,这里也提供了转换的代码
python
#coding=utf-8
import SimpleITK as sitk
def dcm2nii(dcms_path, nii_path):
# 1.构建dicom序列文件阅读器,并执行(即将dicom序列文件"打包整合")
reader = sitk.ImageSeriesReader()
dicom_names = reader.GetGDCMSeriesFileNames(dcms_path)
reader.SetFileNames(dicom_names)
image2 = reader.Execute()
# 2.将整合后的数据转为array,并获取dicom文件基本信息
image_array = sitk.GetArrayFromImage(image2) # z, y, x
origin = image2.GetOrigin() # x, y, z
spacing = image2.GetSpacing() # x, y, z
direction = image2.GetDirection() # x, y, z
# 3.将array转为img,并保存为.nii.gz
image3 = sitk.GetImageFromArray(image_array)
image3.SetSpacing(spacing)
image3.SetDirection(direction)
image3.SetOrigin(origin)
sitk.WriteImage(image3, nii_path)
if __name__ == '__main__':
dcms_path = r'D:\pycharm\deeplearnig\MRIX LUMBAR\Lumbar\Sag T1 Flair - 4' # dicom序列文件所在路径
nii_path = r'.\demo.nii.gz' # 所需.nii.gz文件保存路径
dcm2nii(dcms_path, nii_path)将转换好的nii数据放在images目录下即可
4.2.2 2D推理
代码:
python
from models.sam import SamPredictor, sam_model_registry
from models.sam.modeling.prompt_encoder import attention_fusion
import numpy as np
import os
import torch
import torchvision
import matplotlib.pyplot as plt
from torchvision import transforms
from PIL import Image
import matplotlib.pyplot as plt
from pathlib import Path
from dsc import dice_coeff
import torchio as tio
import nrrd
import PIL
import cfg
from funcs import *
from predict_funs import *
args = cfg.parse_args()
from monai.networks.nets import VNet
args.if_mask_decoder_adapter=True
args.if_encoder_adapter = True
args.decoder_adapt_depth = 2
def predictSlice(image_name, lower_percentile, upper_percentile, slice_id, attention_enabled):
image1_vol = tio.ScalarImage(os.path.join(img_folder, image_name))
print('vol shape: %s vol spacing %s' %(image1_vol.shape,image1_vol.spacing))
image_tensor = image1_vol.data
lower_bound = torch_percentile(image_tensor, lower_percentile)
upper_bound = torch_percentile(image_tensor, upper_percentile)
# Clip the data
image_tensor = torch.clamp(image_tensor, lower_bound, upper_bound)
# Normalize the data to [0, 1]
image_tensor = (image_tensor - lower_bound) / (upper_bound - lower_bound)
image1_vol.set_data(image_tensor)
atten_map= pred_attention(image1_vol,vnet,slice_id,device)
atten_map = torch.unsqueeze(torch.tensor(atten_map),0).float().to(device)
print(atten_map.device)
if attention_enabled:
ori_img,pred_1,voxel_spacing1,Pil_img1,slice_id1 = evaluate_1_volume_withattention(image1_vol,sam_fine_tune,device,slice_id=slice_id,atten_map=atten_map)
else:
ori_img,pred_1,voxel_spacing1,Pil_img1,slice_id1 = evaluate_1_volume_withattention(image1_vol,sam_fine_tune,device,slice_id=slice_id)
mask_pred = ((pred_1>0)==cls).float().cpu()
return ori_img, mask_pred, atten_map
def visualizeSlicePrediction(ori_img, image_name, atten_map, mask_pred):
image = np.rot90(torchvision.transforms.Resize((args.out_size,args.out_size))(ori_img)[0])
image_3d = np.repeat(np.array(image*255,dtype=np.uint8).copy()[:, :, np.newaxis], 3, axis=2)
pred_mask_auto = (mask_pred[0])*255
target_prediction = [103,169,237]
image_pred_auto = drawContour(image_3d.copy(), np.rot90(pred_mask_auto),target_prediction,size=-1,a=0.6)
fig, a = plt.subplots(1,3, figsize=(20,15))
a[0].imshow(image,cmap='gray',vmin=0, vmax=1)
a[0].set_title(image_name)
a[0].axis(False)
a[1].imshow(image_pred_auto,cmap='gray',vmin=0, vmax=255)
a[1].set_title('pre_mask',fontsize=10)
a[1].axis(False)
a[2].imshow(np.rot90(atten_map.cpu()[0]),vmin=0, vmax=1,cmap='coolwarm')
a[2].set_title('atten_map',fontsize=10)
a[2].axis(False)
plt.tight_layout()
plt.show()
device = 'cuda' if torch.cuda.is_available() else 'cpu'
print(device)
checkpoint_directory = './checkpoint' # path to your checkpoint
img_folder = os.path.join('images')
gt_msk_folder = os.path.join('masks')
predicted_msk_folder = os.path.join('predicted_masks')
cls = 1
sam_fine_tune = sam_model_registry["vit_t"](args,checkpoint=os.path.join(checkpoint_directory,'mobile_sam.pt'),num_classes=2)
sam_fine_tune.attention_fusion = attention_fusion()
sam_fine_tune.load_state_dict(torch.load(os.path.join(checkpoint_directory,'bone_sam.pth'),map_location=torch.device(device)), strict = True)
sam_fine_tune = sam_fine_tune.to(device).eval()
vnet = VNet().to(device)
model_directory = './checkpoint'
vnet.load_state_dict(torch.load(os.path.join(model_directory,'atten.pth'),map_location=torch.device(device)))
ori_img, predictedSliceMask, atten_map = predictSlice(
image_name = 'demo.nii.gz',
lower_percentile = 1,
upper_percentile = 99,
slice_id = 3, # slice number
attention_enabled = True, # if you want to use the depth attention
)
visualizeSlicePrediction(
ori_img=ori_img,
image_name='demo.nii.gz',
atten_map=atten_map,
mask_pred=predictedSliceMask,
)结果:
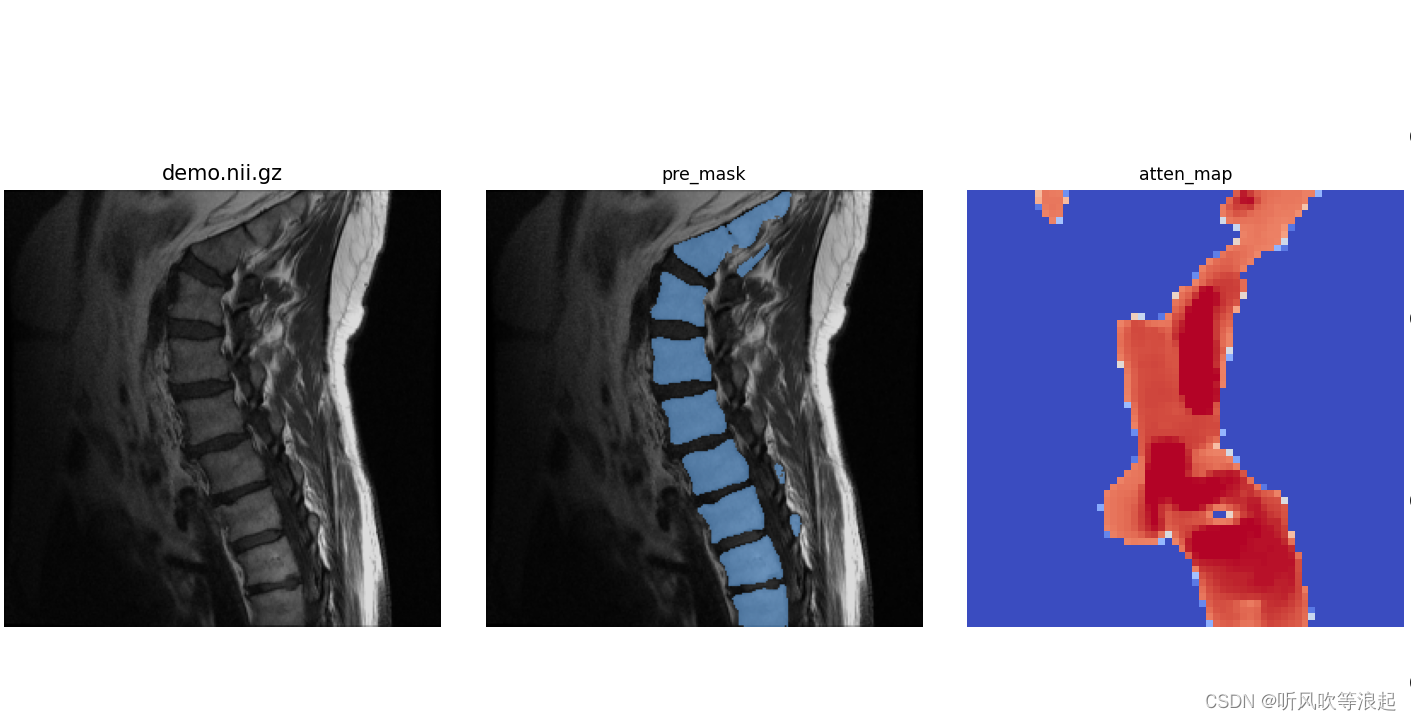
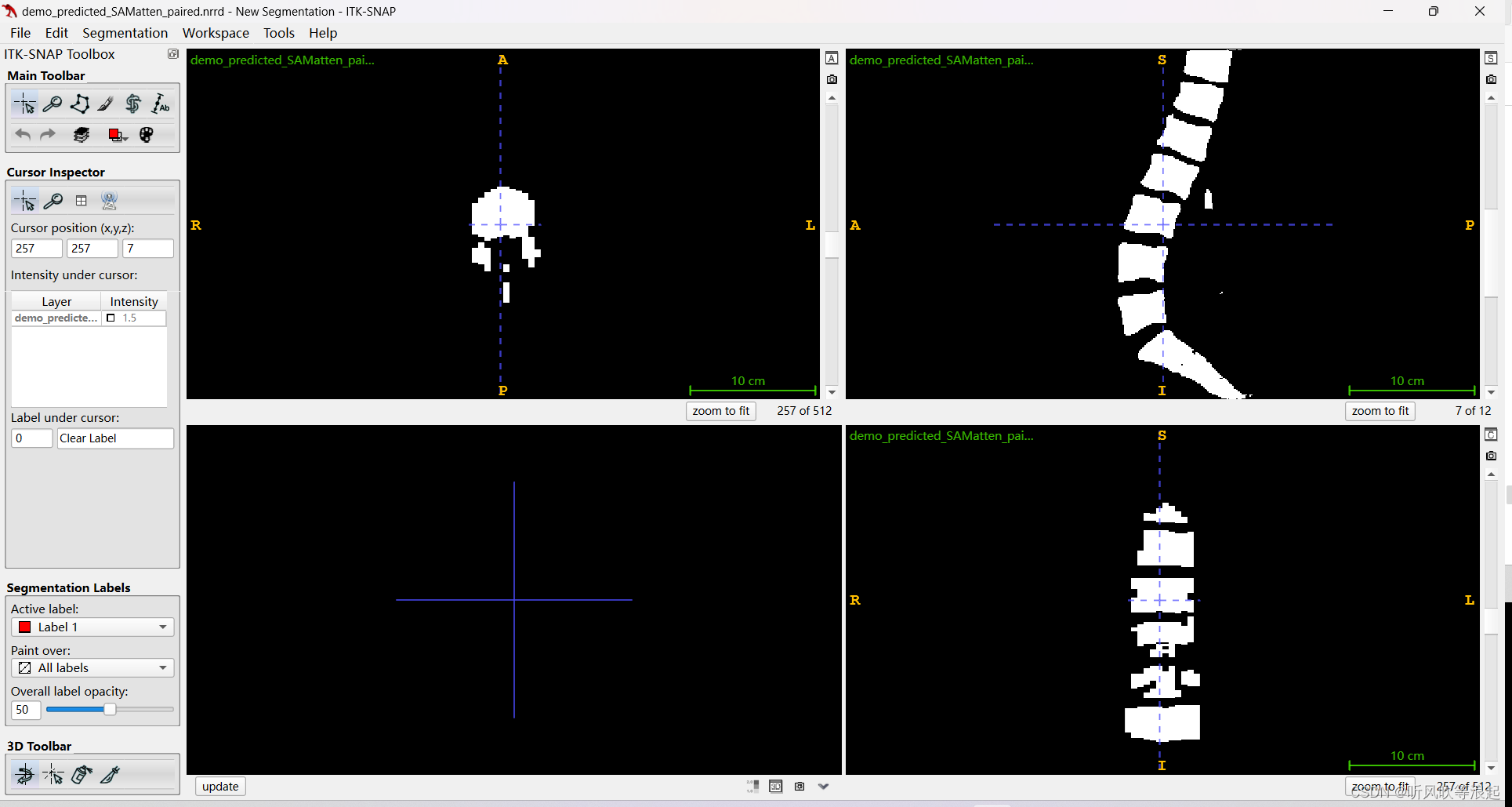
4.2.3 3D推理
代码:
python
from models.sam import SamPredictor, sam_model_registry
from models.sam.modeling.prompt_encoder import attention_fusion
import numpy as np
import os
import torch
import torchvision
import matplotlib.pyplot as plt
from torchvision import transforms
from PIL import Image
import matplotlib.pyplot as plt
from pathlib import Path
from dsc import dice_coeff
import torchio as tio
import nrrd
import PIL
import cfg
from funcs import *
from predict_funs import *
args = cfg.parse_args()
from monai.networks.nets import VNet
args.if_mask_decoder_adapter=True
args.if_encoder_adapter = True
args.decoder_adapt_depth = 2
def predictVolume(image_name, lower_percentile, upper_percentile):
dsc_gt = 0
image1_vol = tio.ScalarImage(os.path.join(img_folder,image_name))
print('vol shape: %s vol spacing %s' %(image1_vol.shape,image1_vol.spacing))
# Define the percentiles
image_tensor = image1_vol.data
lower_bound = torch_percentile(image_tensor, lower_percentile)
upper_bound = torch_percentile(image_tensor, upper_percentile)
# Clip the data
image_tensor = torch.clamp(image_tensor, lower_bound, upper_bound)
# Normalize the data to [0, 1]
image_tensor = (image_tensor - lower_bound) / (upper_bound - lower_bound)
image1_vol.set_data(image_tensor)
mask_vol_numpy = np.zeros(image1_vol.shape)
id_list = list(range(image1_vol.shape[3]))
for id in id_list:
atten_map = pred_attention(image1_vol,vnet,id,device)
atten_map = torch.unsqueeze(torch.tensor(atten_map),0).float().to(device)
ori_img,pred_1,voxel_spacing1,Pil_img1,slice_id1 = evaluate_1_volume_withattention(image1_vol,sam_fine_tune,device,slice_id=id,atten_map=atten_map)
img1_size = Pil_img1.size
mask_pred = ((pred_1>0)==cls).float().cpu()
pil_mask1 = Image.fromarray(np.array(mask_pred[0],dtype=np.uint8),'L').resize(img1_size,resample= PIL.Image.NEAREST)
mask_vol_numpy[0,:,:,id] = np.asarray(pil_mask1)
mask_vol = tio.LabelMap(tensor=torch.tensor(mask_vol_numpy,dtype=torch.int), affine=image1_vol.affine)
mask_save_folder = os.path.join(predicted_msk_folder,'/'.join(image_name.split('/')[:-1]))
Path(mask_save_folder).mkdir(parents=True, exist_ok = True)
mask_vol.save(os.path.join(mask_save_folder,image_name.split('/')[-1].replace('.nii.gz','_predicted_SAMatten_paired.nrrd')))
return mask_vol
device = 'cuda' if torch.cuda.is_available() else 'cpu'
print(device)
checkpoint_directory = './checkpoint' # path to your checkpoint
img_folder = os.path.join('images')
gt_msk_folder = os.path.join('masks')
predicted_msk_folder = os.path.join('predicted_masks')
cls = 1
sam_fine_tune = sam_model_registry["vit_t"](args,checkpoint=os.path.join(checkpoint_directory,'mobile_sam.pt'),num_classes=2)
sam_fine_tune.attention_fusion = attention_fusion()
sam_fine_tune.load_state_dict(torch.load(os.path.join(checkpoint_directory,'bone_sam.pth'),map_location=torch.device(device)), strict = True)
sam_fine_tune = sam_fine_tune.to(device).eval()
vnet = VNet().to(device)
model_directory = './checkpoint'
vnet.load_state_dict(torch.load(os.path.join(model_directory,'atten.pth'),map_location=torch.device(device)))
mask = predictVolume(
image_name = 'demo.nii.gz',
lower_percentile = 1,
upper_percentile = 99
)效果展示: