Title
题目
BrainSegFounder: Towards 3D foundation models for neuroimagesegmentation
BrainSegFounder:迈向用于神经影像分割的3D基础模型
01
文献速递介绍
人工智能(AI)与神经影像分析的融合,特别是多模态磁共振成像(MRI),正在推动脑健康领域的重要进展(Chen等,2022;Segato等,2020;Rao,2023;Owolabi等,2023;Moreno-Blanco等,2019;Rajpurkar等,2022;Khachaturian等,2023)。由于人脑的复杂性,其精密的解剖结构和复杂的功能使神经影像分析面临显著挑战(Moor等,2023;Azad等,2023;Zhang和Metaxas,2024;Segato等,2020;Rajpurkar等,2022)。人工智能在解释复杂神经数据方面的能力有望提高诊断精度,并加深我们对脑部病理的理解。许多研究致力于开发用于特定脑健康分析的人工智能模型,这些研究都在不断丰富神经影像研究领域。
传统上,神经影像人工智能模型需要通过监督学习进行大量的微调,以解决特定的下游任务。诸如nnU-Net(Isensee等,2021)、DeepScan(McKinley等,2019)和DeepMedic(Kamnitsas等,2017)等架构在许多医学计算机视觉挑战中表现出色,例如脑肿瘤分割挑战(BraTS)(Baid等,2021)、医学分割十项全能(MSD)(Antonelli等,2022)以及肿瘤和肝脏自动分割挑战(ATLAS)(Quinton等,2023)。这些进展中的许多都源于在大型无标签数据集上使用自监督预训练方法,将模型编码器和解码器的权重转移到挑战中的较小数据集上(Zhou等,2021;Tang等,2022)。除了这些预训练的改进外,最近还推动了大规模医学数据集的开发(Mei等,2022;Clark等,2013;Bycroft等,2018),以协助这些模型的创建。然而,医学图像分析尚未从最近在自然图像分析和语言处理中的进展中受益,例如"Segment Anything Model(SAM)"(Kirillov等,2023)和LLaMA(Touvron等,2023)模型。
在医学语言处理领域,像MI-Zero(Lu等,2023)和BioViL-T(Bannur等,2023)这样的模型利用对比学习,在医学图像识别中的表征分析和零样本迁移学习方面取得了显著进展。通过利用不同的学习目标,相似的图像-文本对在潜在空间中被拉近,而不相似的对则被推得更远。这类模型推动了组织病理学研究的界限,将基于文本的分析与计算机视觉结合起来。然而,这些模型依赖于训练图像时附带的文本提示(Tiu等,2022)。
Abatract
摘要
The burgeoning field of brain health research increasingly leverages artificial intelligence (AI) to analyze andinterpret neuroimaging data. Medical foundation models have shown promise of superior performance withbetter sample efficiency. This work introduces a novel approach towards creating 3-dimensional (3D) medicalfoundation models for multimodal neuroimage segmentation through self-supervised training. Our approachinvolves a novel two-stage pretraining approach using vision transformers. The first stage encodes anatomicalstructures in generally healthy brains from the large-scale unlabeled neuroimage dataset of multimodal brainmagnetic resonance imaging (MRI) images from 41,400 participants. This stage of pertaining focuses onidentifying key features such as shapes and sizes of different brain structures. The second pretraining stageidentifies disease-specific attributes, such as geometric shapes of tumors and lesions and spatial placementswithin the brain. This dual-phase methodology significantly reduces the extensive data requirements usuallynecessary for AI model training in neuroimage segmentation with the flexibility to adapt to various imagingmodalities. We rigorously evaluate our model, BrainSegFounder, using the Brain Tumor Segmentation (BraTS)challenge and Anatomical Tracings of Lesions After Stroke v2.0 (ATLAS v2.0) datasets. BrainSegFounderdemonstrates a significant performance gain, surpassing the achievements of the previous winning solutionsusing fully supervised learning. Our findings underscore the impact of scaling up both the model complexityand the volume of unlabeled training data derived from generally healthy brains. Both of these factors enhancethe accuracy and predictive capabilities of the model in neuroimage segmentation tasks.
大脑健康研究的快速发展领域越来越多地利用人工智能(AI)来分析和解释神经影像数据。医学基础模型在样本效率上表现出优越的性能,展现了巨大的潜力。本研究提出了一种通过自监督训练创建三维(3D)医学基础模型用于多模态神经影像分割的新方法。我们的方法涉及一种基于视觉转换器(Vision Transformers)的新颖两阶段预训练策略。
第一阶段的预训练编码了大规模无标签神经影像数据集中41,400名参与者的多模态脑部磁共振成像(MRI)图像中的一般健康大脑的解剖结构。该阶段的预训练侧重于识别不同脑结构的关键特征,如形状和大小。第二阶段的预训练识别与疾病相关的特征,如肿瘤和病变的几何形状以及其在大脑中的空间位置。该双阶段方法显著减少了神经影像分割中AI模型训练通常所需的大量数据需求,同时具有适应各种影像模式的灵活性。
我们使用脑肿瘤分割挑战(BraTS)和中风后病变解剖追踪v2.0(ATLAS v2.0)数据集对我们的模型BrainSegFounder进行了严格的评估。BrainSegFounder展现了显著的性能提升,超越了之前使用完全监督学习方法的获胜方案。我们的研究结果强调了增加模型复杂性和从一般健康大脑中衍生的大规模无标签训练数据量对提高神经影像分割任务中模型的准确性和预测能力的重要性。
Background
背景
Method
方法
2.1. Model architecture and pipeline
The BrainSegFounder framework introduces a deep learning training scheme tailored for diverse applications by showcasing a distinctapproach to self-supervised pretraining followed by precise fine-tuning.This section offers a detailed examination of the framework's architecture and its procedural pipeline. It highlights the multi-stageself-supervised pretraining, termed Stage 1 and Stage 2, before proceeding to fine-tuning for downstream tasks. Fig. 1 illustrates BrainSegFounder's architecture. Central to BrainSegFounder is a visiontransformer-based encoder that employs a series of self-attention mechanisms. This encoder is linked with an up-sampling decoder tailored forsegmentation tasks. The architecture is adapted from the SwinUNETRarchitecture (Hatamizadeh et al., 2022) with modified input channelsand input hyperparameters. BrainSegFounder pioneers a novel dualphase self-supervised pretraining method, integrating self-supervisedlearning components within its structure. Stage 1 pretraining exposesthe framework to a wide-ranging dataset of brain MRIs from theUK Biobank dataset, predominantly consisting of healthy individuals.This initial stage equips the model with a thorough comprehension ofstandard brain anatomy, utilizing self-supervised learning to enhanceprediction capabilities. Stage 2 of pretraining advances the model'sproficiency by introducing it to a specialized MRI dataset gearedtowards the downstream task. This phase leverages the architecture'srefined anomaly detection skills, focusing on distinguishing deviationsin brain structure.
2.1. 模型架构和流程
BrainSegFounder框架引入了一种深度学习训练方案,专为多种应用定制,展示了一种独特的自监督预训练方法,随后进行精确的微调。本节详细分析了框架的架构及其流程管道,重点介绍了多阶段自监督预训练(称为阶段1和阶段2),然后进行下游任务的微调。图1展示了BrainSegFounder的架构。
BrainSegFounder的核心是一个基于视觉转换器(Vision Transformer)的编码器,该编码器利用一系列自注意力机制。该编码器与一个针对分割任务的上采样解码器相连。该架构改编自SwinUNETR架构(Hatamizadeh等,2022),并对输入通道和输入超参数进行了修改。BrainSegFounder开创了一种新颖的双阶段自监督预训练方法,在其结构中集成了自监督学习组件。
阶段1预训练让框架接触到来自英国生物样本库(UK Biobank)的大规模脑部MRI数据集,这些数据主要来自健康个体。这个初始阶段使模型对标准脑解剖结构有了透彻的理解,通过自监督学习来增强预测能力。阶段2预训练通过引入专门的MRI数据集,进一步提高模型的能力,以适应下游任务。此阶段利用架构精细化的异常检测能力,重点在于区分脑结构中的偏差。
Results
结果
3.1. Pretraining
The pretraining of our BrainSegFounder models, which varied insize based on the number of parameters, took between 3 to 6 days. Thisprocess utilized a computational setup ranging from 8 to 64 NVIDIAA100 GPUs, each with 80 GB capacity. Fig. 4 illustrates the validationloss during the pretraining phase across different BrainSegFoundermodel sizes.
3.1. 预训练
我们的BrainSegFounder模型的预训练过程根据参数数量的不同,耗时在3至6天之间。该过程使用了从8到64块NVIDIA A100 GPU的计算配置,每块GPU具有80 GB的容量。图4展示了在预训练阶段,不同规模的BrainSegFounder模型的验证损失情况。
Figure
图
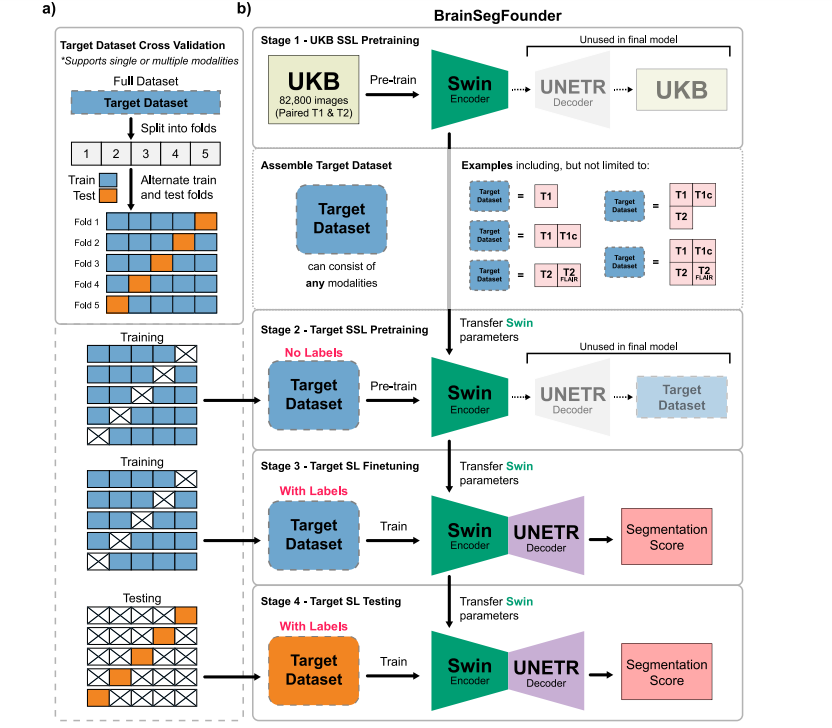
Fig. 1. Overall Study Design. (a) The two-stage pretraining process using Swin Transformer decoders and encoder. Initially, the model is pretrained on the UKB dataset (Stage 1),followed by the downstream task dataset (Stage 2). (b) This is succeeded by fine-tuning on each downstream dataset, with transfer learning applied between each stage.
图1. 整体研究设计。(a) 使用Swin Transformer解码器和编码器的两阶段预训练过程。首先,模型在UKB数据集(阶段1)上进行预训练,然后在下游任务数据集(阶段2)上继续预训练。(b) 随后对每个下游数据集进行微调,并在每个阶段之间应用迁移学习。
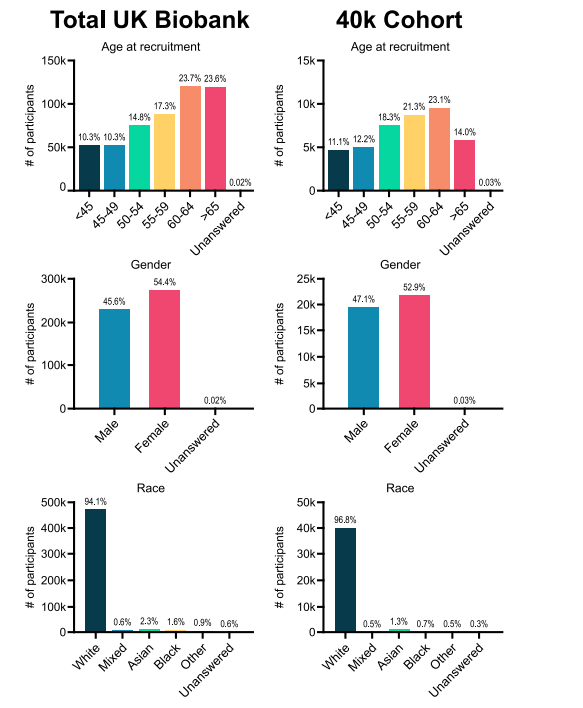
Fig. 2. Visual representation of demographic data from subjects in the UK Biobank in the study
图2. 本研究中来自英国生物样本库受试者的人口统计数据的可视化表示。
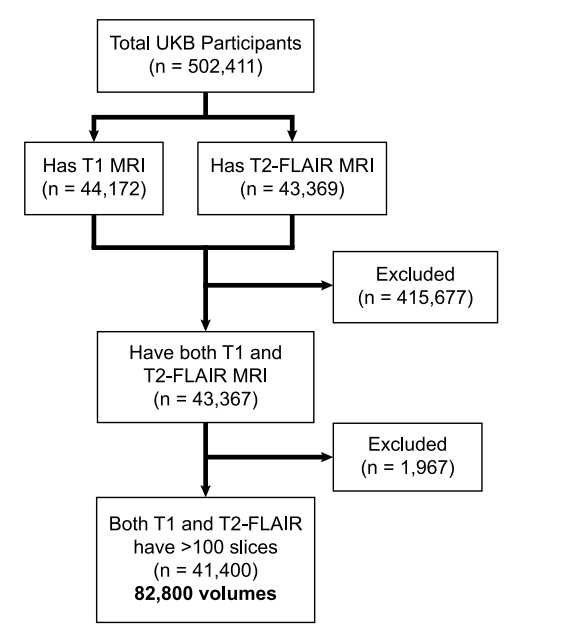
Fig. 3. CONSORT diagram of UKB data used in Stage 1 pretraining
图3. 用于阶段1预训练的英国生物样本库(UKB)数据的CONSORT图。
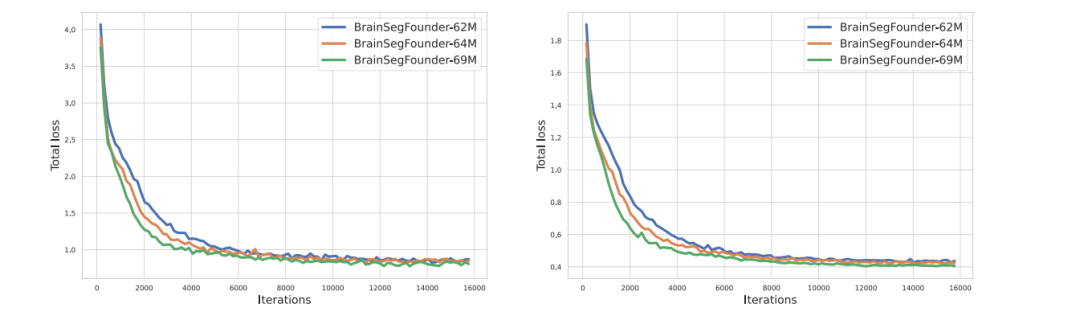
Fig. 4. Training (left) and validation (right) loss of Stage 1-pretraining three different scale of BrainSegFounder models on UKB.
图4. 在英国生物样本库(UKB)上进行阶段1预训练的三种不同规模的BrainSegFounder模型的训练损失(左)和验证损失(右)。
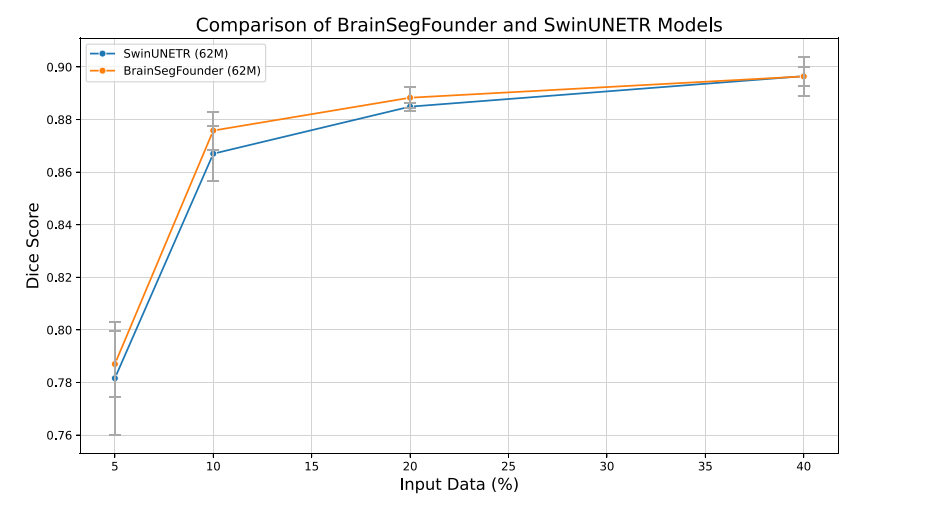
Fig. 5. Dice coefficients for baseline (SwinUNETR) and our model across differentlevels of training data availability. All models were trained 5 times to account forvariability in the input data randomly selected. Error bars represent ± one standarddeviation.
图5. 基线模型(SwinUNETR)和我们的模型在不同训练数据可用性水平下的Dice系数。所有模型均训练了5次,以考虑随机选择的输入数据中的变异性。误差条表示±一个标准差。
Table
表
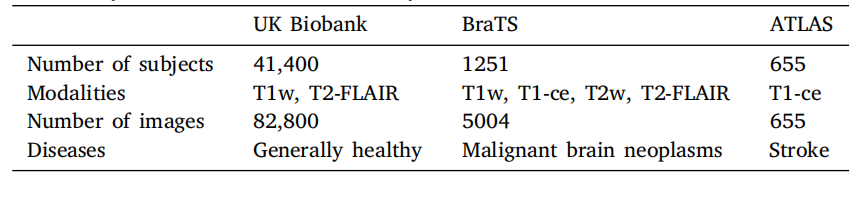
Table 1A summary of the data used in this study.
表1 本研究中使用的数据概述。
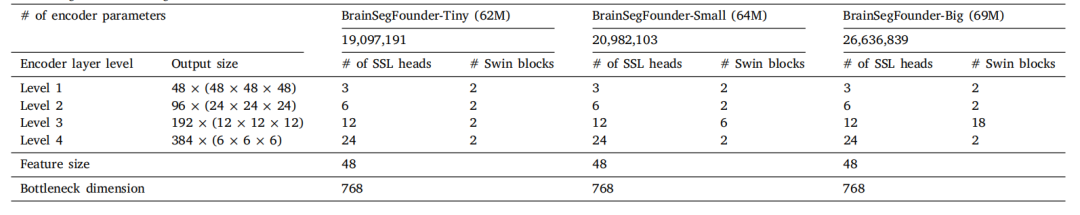
Table 2Pretraining encoder settings.
表2 预训练编码器设置。

Table 3Hardware and training parameters.
表3 硬件和训练参数。
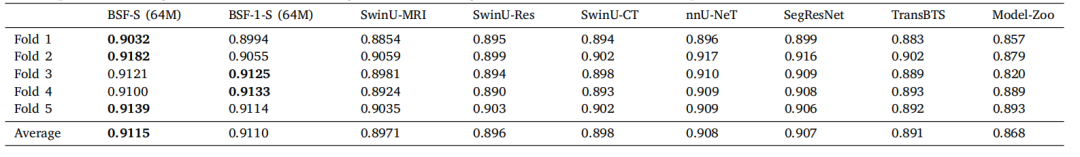
Table 4A comparison of BrainSegFounder (BSF) models' performance in terms of average Dice coefficient on the BraTS challenge. BSF-S indicates our best performing BrainSegFoundermodel (small, 64M parameters). BrainSegFounder models were pretrained with SSL on T1- and T2-weighted MRI 3D volumes and finetuned with supervised learning using allfour modalities present in BraTS. BSF-1-S indicates this model with only the Stage 1 (SSL) pertaining on UKB and without the Stage 2 pretraining step. SwinU models are modelsusing the SwinUNETR architecture trained on BraTS via supervised learning. SwinU-MRI is the model trained directly using supervised learning on BraTS published on GitHub(https://github.com/Project-MONAI/research-contributions/tree/main/SwinUNETR/BRATS21), SwinU-Res is pretrained with SSL on only T1w and T2w and finetuned on BraTS,and SwinU-CT pretrained using CT data and finetuned with supervised learning on BraTS. nnU-Net and SegResNet are former BraTS challenge winners trained using supervisedlearning on our folds. TransBTS is a vision-transformer based segmentation algorithm optimized for brain segmentation. Model-Zoo is a bundle of models published by MONAIthat can perform BraTS segmentation out of the box using their ''Brats mri segmentation'' [sic] model found at https://monai.io/model-zoo.html.
表4 BrainSegFounder(BSF)模型在BraTS挑战中的平均Dice系数表现比较。BSF-S表示我们表现最佳的BrainSegFounder模型(小型,64M参数)。BrainSegFounder模型通过自监督学习(SSL)在T1和T2加权的MRI三维体积上进行预训练,并使用BraTS中所有四种模态的监督学习进行微调。BSF-1-S表示仅在UKB上进行阶段1(SSL)预训练且未进行阶段2预训练的模型。SwinU模型是使用SwinUNETR架构通过监督学习在BraTS上训练的模型。SwinU-MRI是在BraTS上直接使用监督学习训练的模型,发布在GitHub上(链接)。SwinU-Res模型仅在T1w和T2w数据上使用SSL预训练,然后在BraTS上进行微调。SwinU-CT模型使用CT数据预训练,并在BraTS上通过监督学习进行微调。nnU-Net和SegResNet是先前BraTS挑战的获胜者,使用监督学习在我们的数据集上训练。TransBTS是一种基于视觉转换器的脑分割优化算法。Model-Zoo是MONAI发布的一组模型,能够直接使用其"Brats mri segmentation"模型(可在此处找到)执行BraTS分割任务。
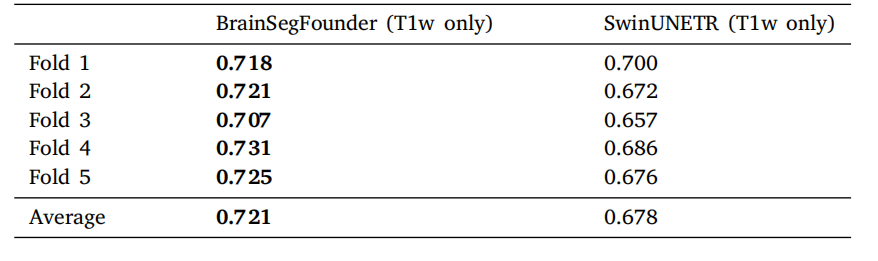
Table 5Performance comparison of modality restricted models on the BraTS dataset. SwinUNETR is fully supervised learning on T1-weighted MRI without pretraining, whileBrainSegFounder uses our multi-stage pretraining on UKB and BraTS T1-weighted MRIand is then finetuned on BraTS T1-weighted MRI.
表5 模态受限模型在BraTS数据集上的性能比较。SwinUNETR是在没有预训练的情况下对T1加权MRI进行完全监督学习,而BrainSegFounder使用我们的多阶段预训练(在UKB和BraTS T1加权MRI上)后,在BraTS T1加权MRI上进行微调。
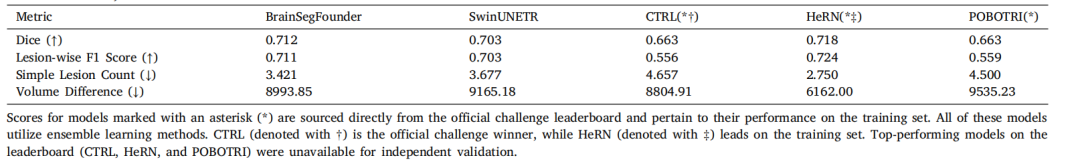
Table 6Performance comparison of segmentation models on the ATLAS v2.0 dataset. All metrics from the challenge (Dice coefficient, Lesion-wise F1 Score, Simple Lesion Count, andVolume Difference) are included for each model.
表6 分割模型在ATLAS v2.0数据集上的性能比较。每个模型的所有挑战指标(Dice系数、病灶级F1得分、简单病灶计数和体积差异)均包括在内。
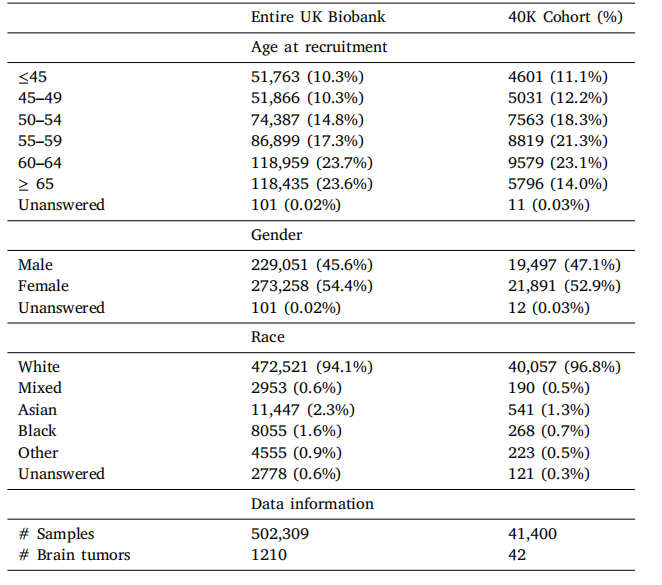
Table A.7UKB Data Demographic information.
表A.7 UKB数据的人口统计信息。
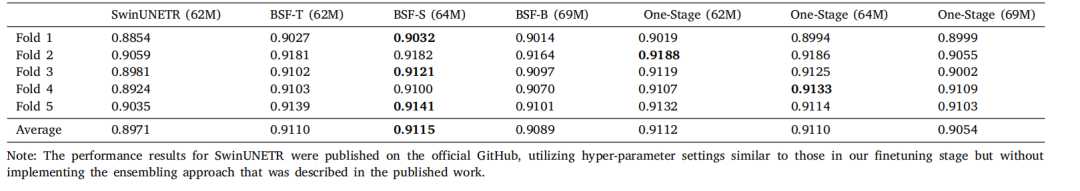
Table A.8Comparison of BrainSegFounder models through 5-fold cross-validation with metric Dice coefficient on BraTS. SwinUNETR is the winning solution on BraTS challenge 2021,which is performed with fully supervised learning without UKB pretraining. BrainSegFounder is the proposed method, which is conducted with the two-stage pretraining and thenfinetuning on the target dataset. The one-stage means that pretraining on UKB is performed but not on the BraTS.
表A.8 通过5折交叉验证对BrainSegFounder模型在BraTS上的Dice系数进行比较。SwinUNETR是2021年BraTS挑战赛的获胜方案,使用完全监督学习进行训练,没有UKB预训练。BrainSegFounder是提出的方法,采用了两阶段预训练,随后在目标数据集上进行微调。"一阶段"意味着仅在UKB上进行预训练,而不在BraTS上进行。
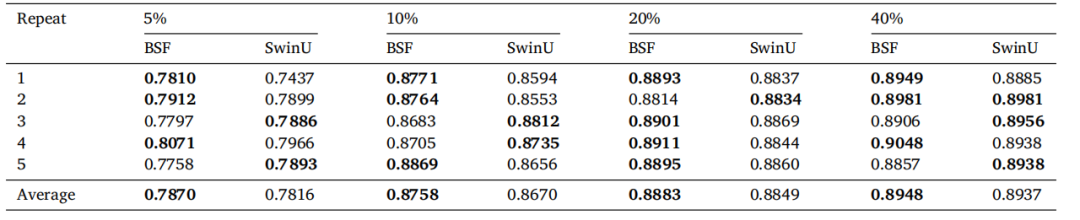
Table A.9Comparison of BrainSegFounder (BSF) and SwinUNETR (SwinU) Baseline models trained on 5 repeats of varying percentages of the input data.Data was randomly sampled from the BraTS training dataset, and models were evaluated on the testing dataset
表A.9 BrainSegFounder(BSF)和SwinUNETR(SwinU)基线模型在不同百分比的输入数据上进行5次重复训练的比较。数据从BraTS训练数据集中随机抽样,模型在测试数据集上进行评估。