01 3D genome data analysis guides
https://trackc.readthedocs.io/en/latest/install.html #官网
https://github.com/seqyuan/trackc #官网
https://trackc.readthedocs.io/en/latest/analysis_guide/index.html #HiC可视化案例GridSpec
在基因组区域中可视化多组学数据通常需要结合多个轨迹(tracks)。为了创建适合发表的高质量图像,精细调整每个轨迹的比例非常重要。快速调整轨迹比例以及轻松添加或移除轨迹的能力是一个重要问题。
trackc 提供了两种方法来绘制布局:
-
trackc.tenon -
trackc.make_spec
这些函数为用户提供了一种快速灵活的方法,用于调整每个轨迹的比例,同时方便地在多组学可视化布局中添加或移除轨迹。
1. trackc.tenon
-
trackc.tenon返回一个对象。例如:ten = trackc.tenon(width=5, height=1)在此示例中,
ten是一个表示虚拟图形的对象,其宽度为 5,高度为 1。用户可以通过以下方法向该图形添加轨迹:ten.add(pos='bottom', height=4, hspace=0.1)-
height参数 表示新添加轨迹的相对高度。例如,如果虚拟图形的高度为 1,而新轨迹的高度为 4,则子图的实际高度将为 1 * 4。 -
使用
ten.add方法 类似于搭建积木,用户可以通过pos参数 选择将新轨迹添加到顶部(top)或底部(bottom)。 -
hspace=0.1参数 控制新轨迹与相邻轨迹之间的间距。
-
2. trackc.make_spec
trackc.make_spec定义整个图像的大小,允许用户从上到下或从左到右设置一组子图,可以通过height_ratios或width_ratios控制各部分的比例。
可用轨迹
本节包括多个快速教程,展示了使用 trackc 进行组学数据可视化的功能。
- GridSpec
- scale
- mapC
- mapc_markline
- Virtual4C
- gene
- bigwig
- bed
- bedGraphMatrix
- links
- zoomin
- highlight line
02 安装
trackc 运行需要 Python 版本 >= 3.8。
通过 PyPI 安装
使用以下命令安装 trackc:
pip install trackc
更新 trackc 的命令:
pip install --upgrade trackc
指定版本和安装源的命令:
pip install -i https://pypi.org/simple trackc==版本号开发版本
从 GitHub 安装 trackc 的命令:
pip install git+https://github.com/seqyuan/trackc@main03 heatmap
conf-hicmap_1.yml等很多trackc:
- ax: t1
height: 2
track_type: hicmap
track_para:
mat:
method: extractContactRegions
clr: /path/GSM4417639_SK-N-DZ_C6BC81F2_b38d5.mcool::/resolutions/5120000
row_regions:
- chr6
- chr8
mapC:
cmap: plasma运行trackc cli conf-hicmap_1.yml -s 4,1 -o chr6_chr8.pdf
conf-hicmap_4.yml
trackc:
- ax: t1
height: 1
track_type: hicmap
track_para:
mat:
method: extractCisContact
clr: ./github/seqyuan/trackc_data/examples/HPDE6C7.chr18.mcool::/resolutions/25000
mapC:
map_type: tri
height: 50
label: tri
- ax: t2
height: 1
hspace: 0.3
track_type: hicmap
track_para:
mat2:
method: extractCisContact
clr: ./github/seqyuan/trackc_data/examples/HPDE6C7.chr18.mcool::/resolutions/25000
mapC:
map_type: tri
height: 50
label: tri-mat2
- ax: t3
height: 2
hspace: 0.3
track_type: hicmap
track_para:
mat:
method: extractCisContact
clr: ./github/seqyuan/trackc_data/examples/HPDE6C7.chr18.mcool::/resolutions/25000
mapC:
map_type: tri
height: 50
label: tri-symmetric
symmetric: True
- ax: t4
height: 1
hspace: 0.3
track_type: hicmap
track_para:
mat:
method: extractCisContact
clr:./github/seqyuan/trackc_data/examples/BxPC3.chr18.mcool::/resolutions/25000
extend: 50
mapC:
cmap: PuBu
map_type: rec
height: 50
label: rec
- ax: t5
height: 1
hspace: 0.3
track_type: hicmap
track_para:
mat2:
method: extractCisContact
clr: ./github/seqyuan/trackc_data/examples/BxPC3.chr18.mcool::/resolutions/25000
extend: 50
mapC:
cmap: cividis
map_type: rec
height: 50
label: rec-mat2
- ax: t6
height: 2
hspace: 0.3
track_type: hicmap
track_para:
mat2:
method: extractCisContact
clr: ./github/seqyuan/trackc_data/examples/BxPC3.chr18.mcool::/resolutions/25000
extend: 50
mapC:
cmap: magma
map_type: rec
height: 50
symmetric: True
logdata: True
label: rec-symmetric
- ax: t1
track_type: scale_track
track_para:
scale_adjust: Mb
tick_pos: top
ratio2ax: 1运行
trackc cli conf-hicmap_4.yml -r 18:10000000-14500000
04 rearranged_interactions
ectopic_interactions.yaml
trackc:
- ax: t1
height: 2
track_type: hicmap
track_para:
mat:
method: extractContactRegions
clr: ./github/seqyuan/trackc_data/examples/BxPC3.chr18.mcool::/resolutions/50000
balance: False
row_regions: 18:45000000-78077248
mapC:
map_type: tri
label: tumor res=50k
cmap: PuBu
maxrange: 70
ax_on: False
- ax: t1
track_type: scale_track
track_para:
region: 18:45000000-78077248
scale_adjust: Mb
tick_pos: top
ratio2ax: 0.3
space: 0.01
tick_fontsize: 6
label_fontsize: 7
- ax: t2
height: 0.2
track_type: zoomin
track_para:
raw_regions: 18:45000000-78077248
zoomin_regions:
- 18:47340000-50370000
- 18:61140000-63630000
- 18:74030000-77560000
line_on: False
fill: True
alpha: 0.5
- ax: t3
height: 0.4
track_type: multi_scale_track
track_para:
regions:
- 18:47340000-50370000
- 18:61140000-63630000
- 18:74030000-77560000
scale_adjust: Mb
intervals: 2
tick_fontsize: 7
- ax: t4
height: 2
track_type: hicmap
track_para:
mat2:
method: extractContactRegions
clr: ./github/seqyuan/trackc_data/examples/BxPC3.chr18.mcool::/resolutions/25000
balance: False
row_regions:
- 18:47340000-50370000
- 18:61140000-63630000
- 18:74030000-77560000
mapC:
map_type: tri
label: ectopic interactions
cmap: PuBu
ax_on: False
maxrange: 100
- ax: t5
height: 0.2
track_type: zoomin
track_para:
raw_regions:
- 18:47340000-50370000
- 18:61140000-63630000
- 18:74030000-77560000
zoomin_regions:
- 18:47400000-48280000
- 18:75280000-74030000
line_on: False
fill: True
alpha: 0.5
- ax: t6
height: 0.4
track_type: multi_scale_track
track_para:
regions:
- 18:47400000-48280000
- 18:75280000-74030000
scale_adjust: Mb
intervals: 2
tick_fontsize: 7
- ax: t7
height: 1.25
track_type: hicmap
track_para:
mat:
method: extractContactRegions
clr: ./github/seqyuan/trackc_data/examples/BxPC3.chr18.mcool::/resolutions/25000
balance: False
row_regions:
- 18:47400000-48280000
- 18:75280000-74030000
mapC:
map_type: tri
label: neo tad
cmap: PuBu
height: 40
ax_on: False
maxrange: 200
minrange: 10
- ax: t8
height: 0.5
track_type: bw_track
track_para:
bw: ./github/seqyuan/trackc_data/examples/GSM3178671_BxPC3_H3K27ac.bigwig
regions:
- 18:47400000-48280000
- 18:75280000-74030000
label: H3K27ac
binsize: 2000
color:
- '#5B7695'
- ax: t9
height: 2.8
track_type: gene_track
track_para:
bed12: ./github/seqyuan/trackc_data/examples/hg19_chr18.gene.bed12
regions:
- 18:47400000-48280000
- 18:75280000-74030000
line: 12
gene_fontszie: 8运行
trackc cli ectopic_interactions.yaml -s 5,1
05 Virtual 4C
4C.yaml
trackc:
- ax: t1
height: 4
track_type: hicmap
track_para:
mat:
method: extractContactRegions
clr: ./github/seqyuan/trackc_data/tutorials/4C/GSM4604287_1360.sub.cool
balance: False
mapC:
map_type: tri
label: AML 1360
cmap: PuBu
maxrange: 6
minrange: -6
- ax: t2
height: 0.7
track_type: gene_track
track_para:
bed12: ./github/seqyuan/trackc_data/tutorials/4C/GRCh38.gene.chr8chr14.bed12
line: 3
gene_fontszie: 10
show_label:
- MYC
- ax: t3
height: 0.7
track_type: bw_track
track_para:
bw: ./github/seqyuan/trackc_data/tutorials/4C/GSM4604189_H3K27ac.bw
maxrange: 10
label: H3K27ac
binsize: 10000
invert_y: False
- ax: t4
height: 0.6
track_type: virtual4C
track_para:
clr: ./github/seqyuan/trackc_data/tutorials/4C/GSM4604287_1360.sub.cool
maxrange: 10
label: Virtual 4C
target_color: tab:red
track_type: line
target: chr8:127735434-127735435
- ax: t5
height: 0.6
track_type: multi_scale_track
track_para:
scale_adjust: Mb
intervals: 1
tick_fontsize: 8运行
trackc cli 4C.yaml -s 6,0.75 -r 'chr8:127000000-129200000 chr14:96500000-99300000'
06 常用命令行
import trackc as tc
ten = tc.tenon(figsize=(8,1))
ten.add(pos='bottom', height=3.6)
ten.add(pos='bottom', height=1)
chr6_len = clr.chromsizes['chr6']
chr8_len = clr.chromsizes['chr8']
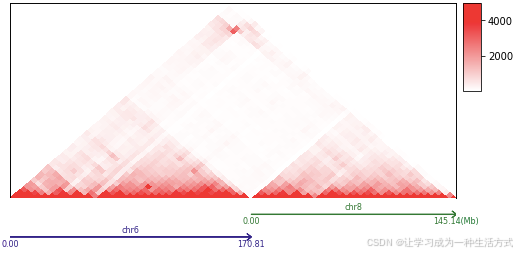
mat = tc.tl.extractContactRegions(clr='GSM4417639_SK-N-DZ_C6BC81F2_b38d5.mcool::/resolutions/5120000', row_regions=['chr6', 'chr8'])
tc.pl.mapC(ax=ten.axs(0), mat=mat.cmat, map_type='triangle', maxrange=5000)
tc.pl.multi_scale_track(ax=ten.axs(1), regions=['chr6:0-{0}'.format(chr6_len), 'chr8:0-{0}'.format(chr8_len)],
scale_adjust='Mb', intervals=2, tick_rotation=0)
# tc.savefig('hicmap.pdf')