介绍
不同期刊配色大多数时候不一样,为了更好符合期刊图片颜色的配色,有人开发了ggsci这个R包。它提供以下函数:
-
scale_color_palname()
-
scale_fill_palname()
对应不同期刊的color和fill函数。
导入数据+R包
R
library("ggsci")
library("ggplot2")
library("gridExtra")
data("diamonds")
p1 <- ggplot(subset(diamonds, carat >= 2.2),
aes(x = table, y = price, colour = cut)) +
geom_point(alpha = 0.7) +
geom_smooth(method = "loess", alpha = 0.05, size = 1, span = 1) +
theme_bw()
p2 <- ggplot(subset(diamonds, carat > 2.2 & depth > 55 & depth < 70),
aes(x = depth, fill = cut)) +
geom_histogram(colour = "black", binwidth = 1, position = "dodge") +
theme_bw()NPG: Nature Publishing Group
R
p1_npg <- p1 + scale_color_npg()
p2_npg <- p2 + scale_fill_npg()
grid.arrange(p1_npg, p2_npg, ncol = 2)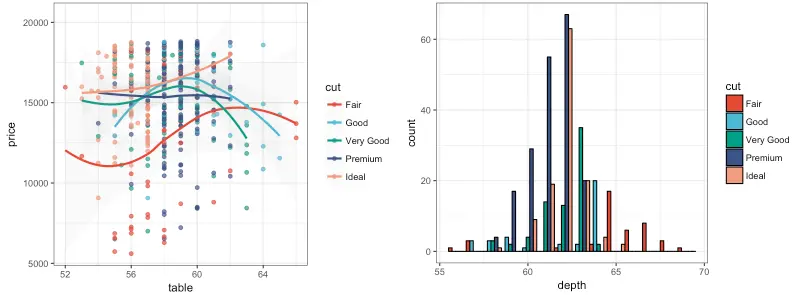
AAAS: American Association for the Advancement of Science
R
p1_aaas <- p1 + scale_color_aaas()
p2_aaas <- p2 + scale_fill_aaas()
grid.arrange(p1_aaas, p2_aaas, ncol = 2)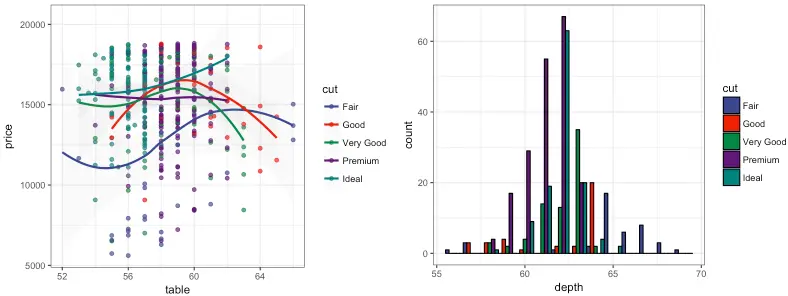
NEJM:The New England Journal of Medicine
R
p1_nejm <- p1 + scale_color_nejm()
p2_nejm <- p2 + scale_fill_nejm()
grid.arrange(p1_nejm, p2_nejm, ncol = 2)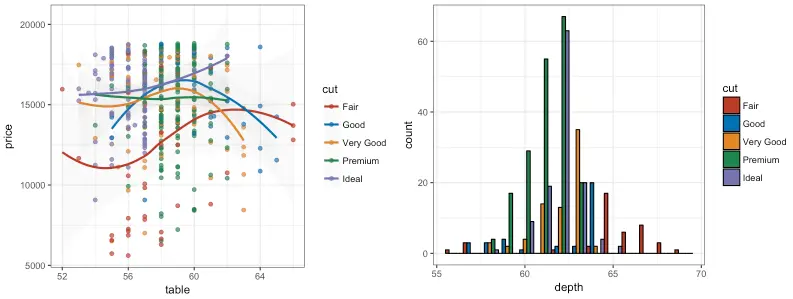
Lancet: Lancet journals
R
p1_lancet <- p1 + scale_color_lancet()
p2_lancet <- p2 + scale_fill_lancet()
grid.arrange(p1_lancet, p2_lancet, ncol = 2)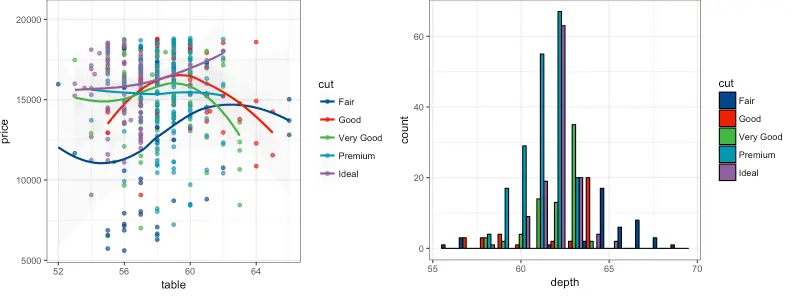
JAMA: The Journal of the American Medical Association
R
p1_jama <- p1 + scale_color_jama()
p2_jama <- p2 + scale_fill_jama()
grid.arrange(p1_jama, p2_jama, ncol = 2)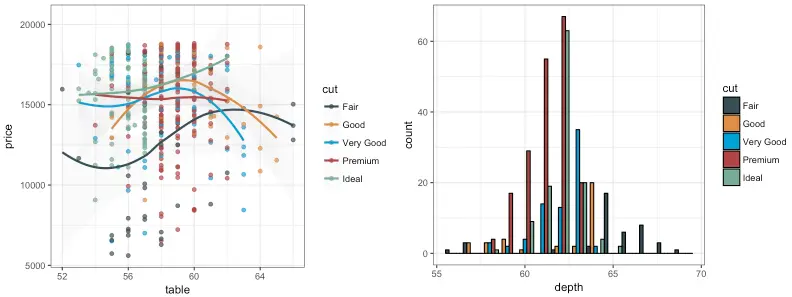
UCSCGB: UCSC Genome Browser
R
p1_ucscgb <- p1 + scale_color_ucscgb()
p2_ucscgb <- p2 + scale_fill_ucscgb()
grid.arrange(p1_ucscgb, p2_ucscgb, ncol = 2)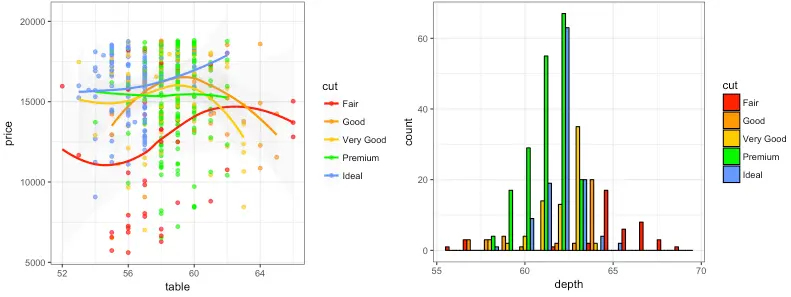
Tron Legacy
R
p1_tron <- p1 + theme_dark() + theme(
panel.background = element_rect(fill = "#2D2D2D"),
legend.key = element_rect(fill = "#2D2D2D")) +
scale_color_tron()
p2_tron <- p2 + theme_dark() + theme(
panel.background = element_rect(fill = "#2D2D2D")) +
scale_fill_tron()
grid.arrange(p1_tron, p2_tron, ncol = 2)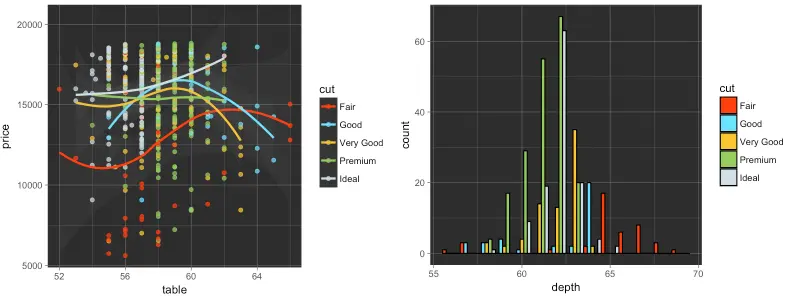
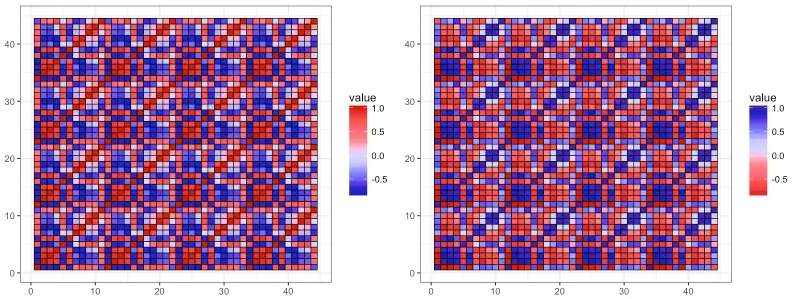
GSEA: GSEA GenePattern
R
library("reshape2")
data("mtcars")
cor <- cor(unname(cbind(mtcars, mtcars, mtcars, mtcars)))
cor_melt <- melt(cor)
p3 <- ggplot(cor_melt, aes(x = Var1, y = Var2, fill = value)) +
geom_tile(colour = "black", size = 0.3) +
theme_bw() +
theme(axis.title.x = element_blank(),
axis.title.y = element_blank())
p3_gsea <- p3 + scale_fill_gsea()
p3_gsea_inv <- p3 + scale_fill_gsea(reverse = TRUE)
grid.arrange(p3_gsea, p3_gsea_inv, ncol = 2)