✨作者主页 :IT研究室✨
个人简介:曾从事计算机专业培训教学,擅长Java、Python、微信小程序、Golang、安卓Android等项目实战。接项目定制开发、代码讲解、答辩教学、文档编写、降重等。
文章目录
一、前言
系统介绍
基于大数据的宫颈癌风险因素分析与可视化系统是一个集数据存储、分析和可视化于一体的综合性医疗数据分析平台。该系统采用Hadoop生态圈作为分布式存储和计算的基础架构,通过HDFS分布式文件系统管理大规模医疗数据,利用Spark和Spark SQL进行高效的数据处理和分析。系统后端采用Django框架构建RESTful API服务,前端基于Vue框架开发响应式用户界面,集成ElementUI组件库提供优质的用户体验,通过Echarts图表库实现丰富的数据可视化效果。系统支持多维度的宫颈癌风险因素分析,包括患者年龄分布、性行为史、STDs感染情况、避孕方式、吸烟史等关键指标的统计分析,能够进行聚类分析识别高危人群,提供Hinselmann、Schiller、Citology等多种筛查方法的验证对比分析。同时系统构建了综合性的数据可视化大屏,通过多种图表形式直观展示风险因素分布规律,为医疗机构的临床决策和预防工作提供数据支撑。
选题背景
宫颈癌作为女性常见的恶性肿瘤之一,在全球范围内对女性健康构成重大威胁。当前医疗机构在宫颈癌预防和筛查过程中积累了大量的患者数据,包括人口学特征、生活习惯、性行为史、病毒感染情况等多维度信息,但这些数据往往分散存储,缺乏有效的整合分析。传统的数据处理方式已难以满足海量医疗数据的分析需求,医生和研究人员难以从复杂的数据中快速识别出关键的风险因素和规律。与此同时,现有的宫颈癌风险评估主要依赖于医生的临床经验和单一检查结果,缺乏基于大数据的综合性分析工具。随着大数据技术和机器学习算法的发展成熟,为医疗数据的深度挖掘和智能分析提供了新的技术手段。医疗机构迫切需要一个能够处理大规模数据、进行多维度分析、提供直观可视化展示的系统平台,来辅助医生进行更精准的风险评估和预防决策。
选题意义
本研究的实际意义主要体现在为医疗机构提供一个相对完整的数据分析工具,虽然作为毕业设计项目在功能深度上存在一定局限,但在数据整合和可视化方面能够起到一定的辅助作用。系统通过整合患者的多维度信息,可以帮助医生更全面地了解风险因素的分布规律,在一定程度上提升风险评估的效率。对于医院信息化建设而言,该系统提供了一个可参考的技术实现方案,展示了大数据技术在医疗数据分析中的应用潜力。从技术层面来看,本项目探索了Hadoop和Spark等大数据技术在医疗领域的具体应用,为类似的医疗数据分析项目提供了一定的技术参考。同时,系统设计的可视化界面能够让医疗工作者更直观地观察数据趋势,在日常的数据查看和初步分析中发挥一些作用。虽然本系统在算法复杂度和数据规模处理能力上还有提升空间,但作为一个初步的技术实践,它验证了将传统医疗数据与现代大数据技术结合的可行性,为后续更深入的研究和开发奠定了基础。
二、开发环境
- 大数据框架:Hadoop+Spark(本次没用Hive,支持定制)
- 开发语言:Python+Java(两个版本都支持)
- 后端框架:Django+Spring Boot(Spring+SpringMVC+Mybatis)(两个版本都支持)
- 前端:Vue+ElementUI+Echarts+HTML+CSS+JavaScript+jQuery
- 详细技术点:Hadoop、HDFS、Spark、Spark SQL、Pandas、NumPy
- 数据库:MySQL
三、系统界面展示
- 基于大数据的宫颈癌风险因素分析与可视化系统界面展示:
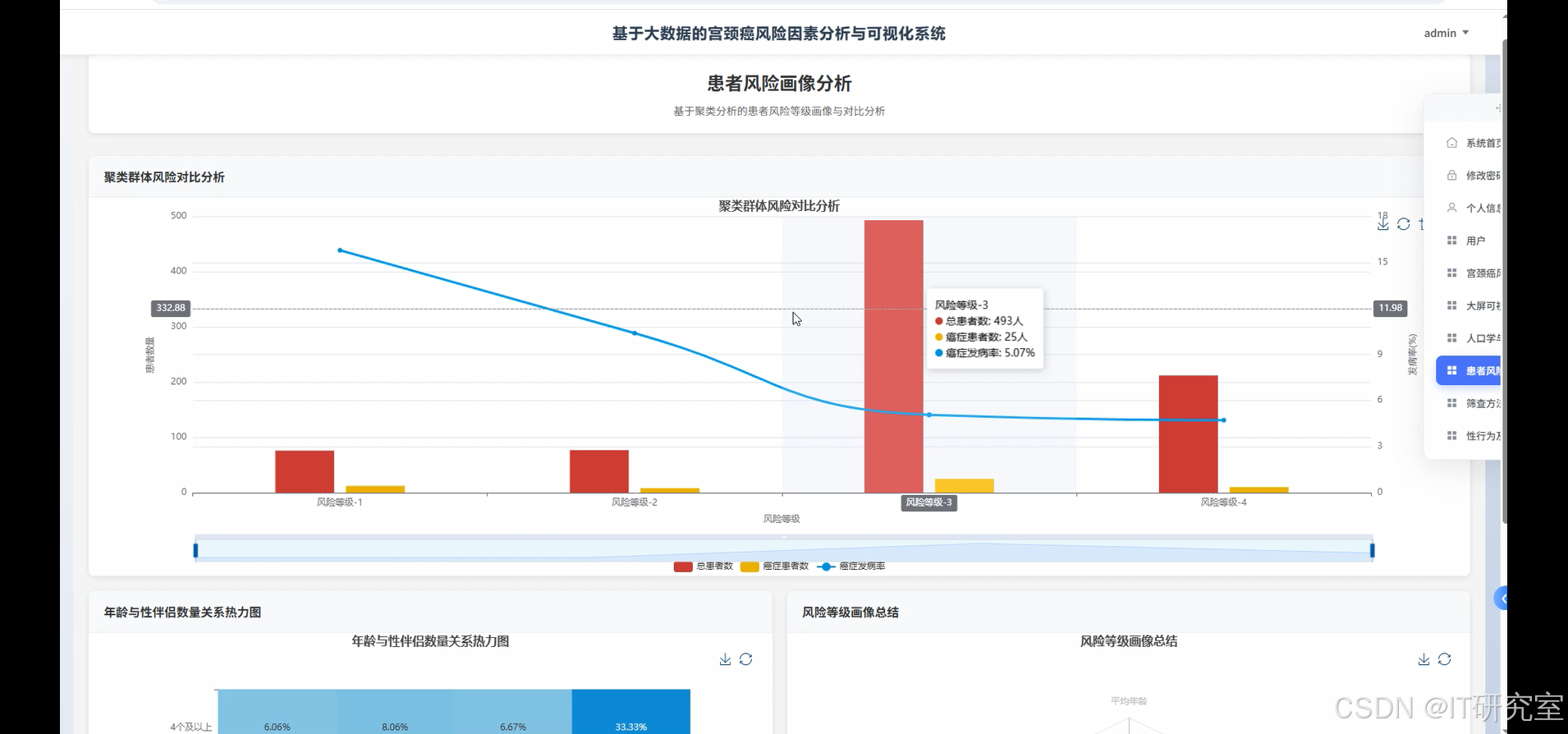
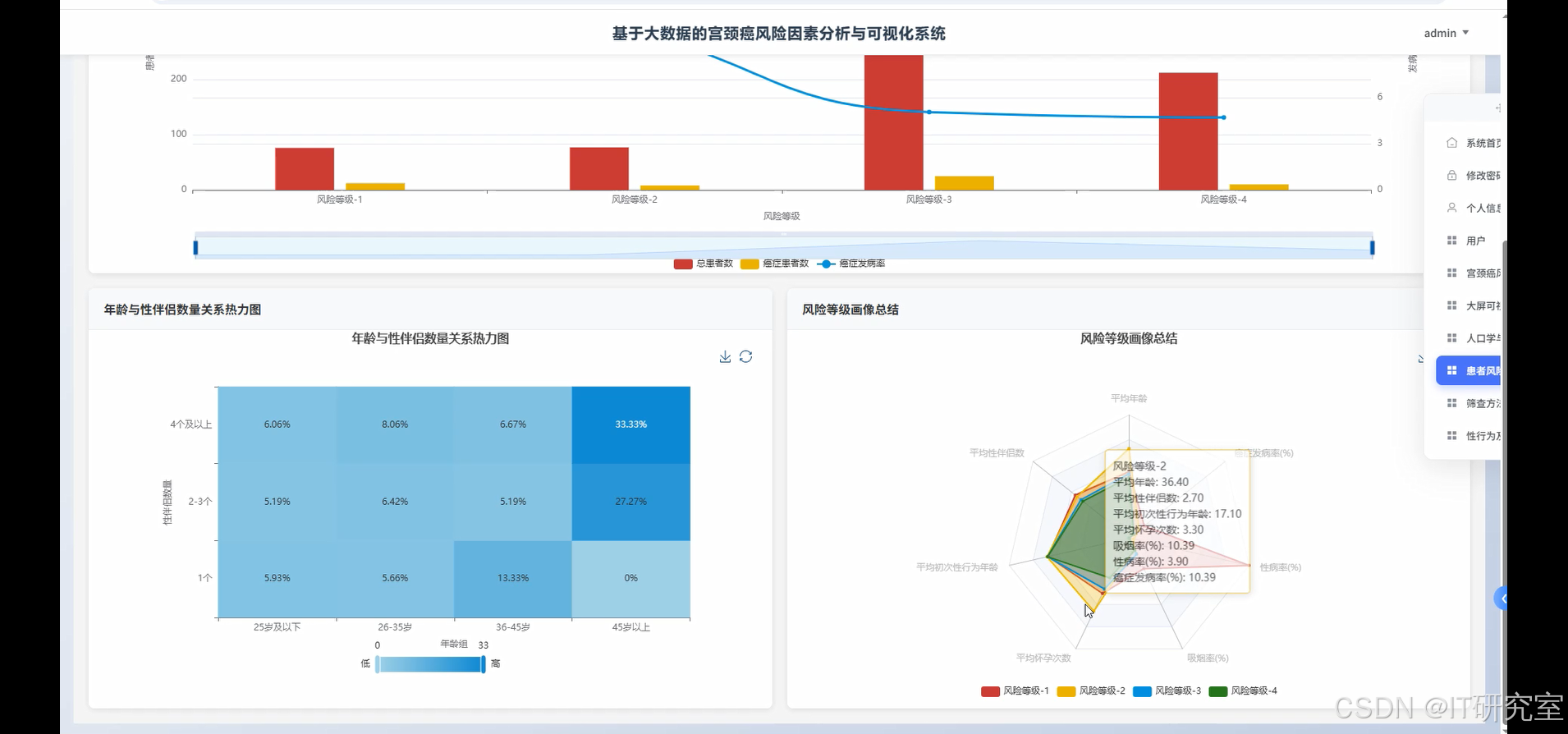
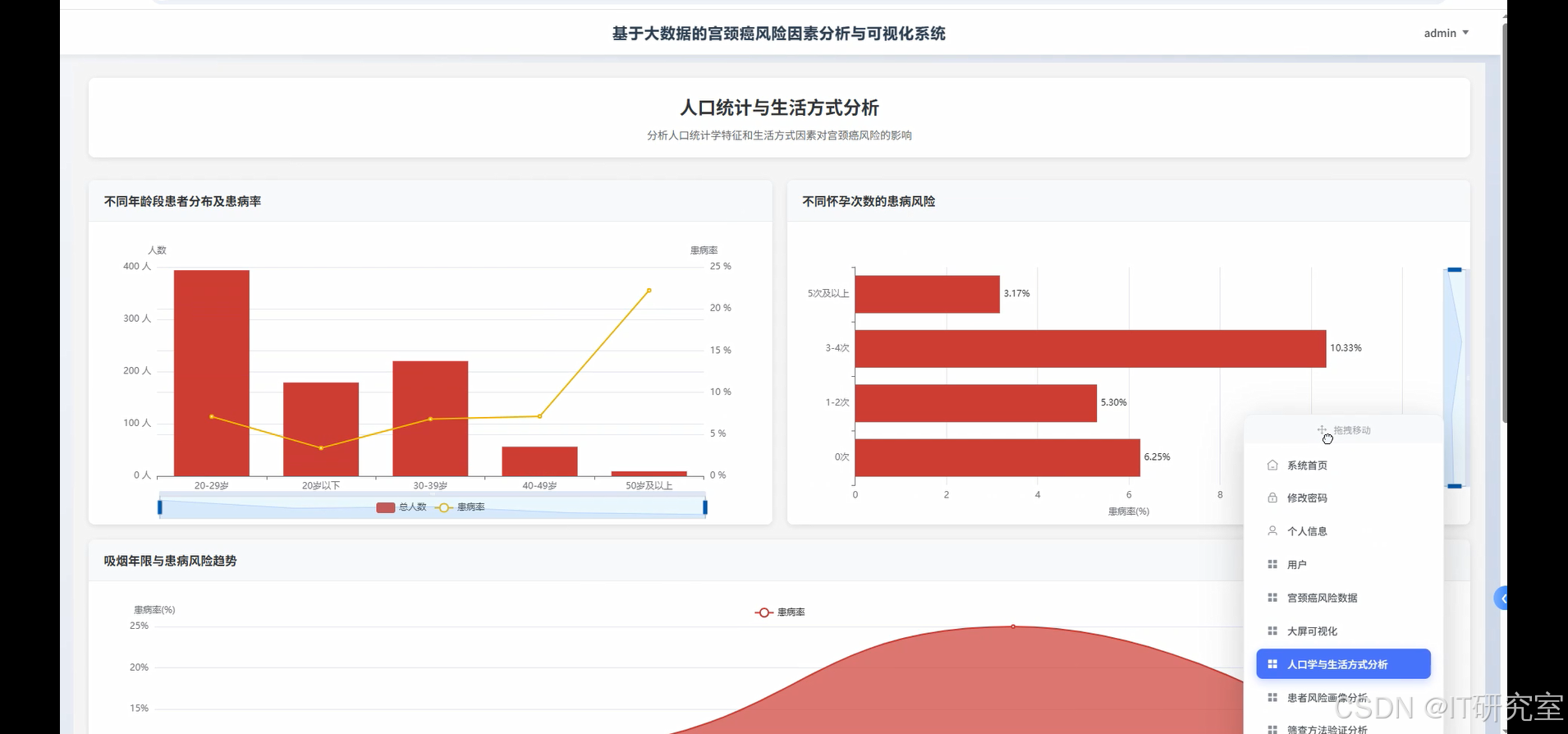
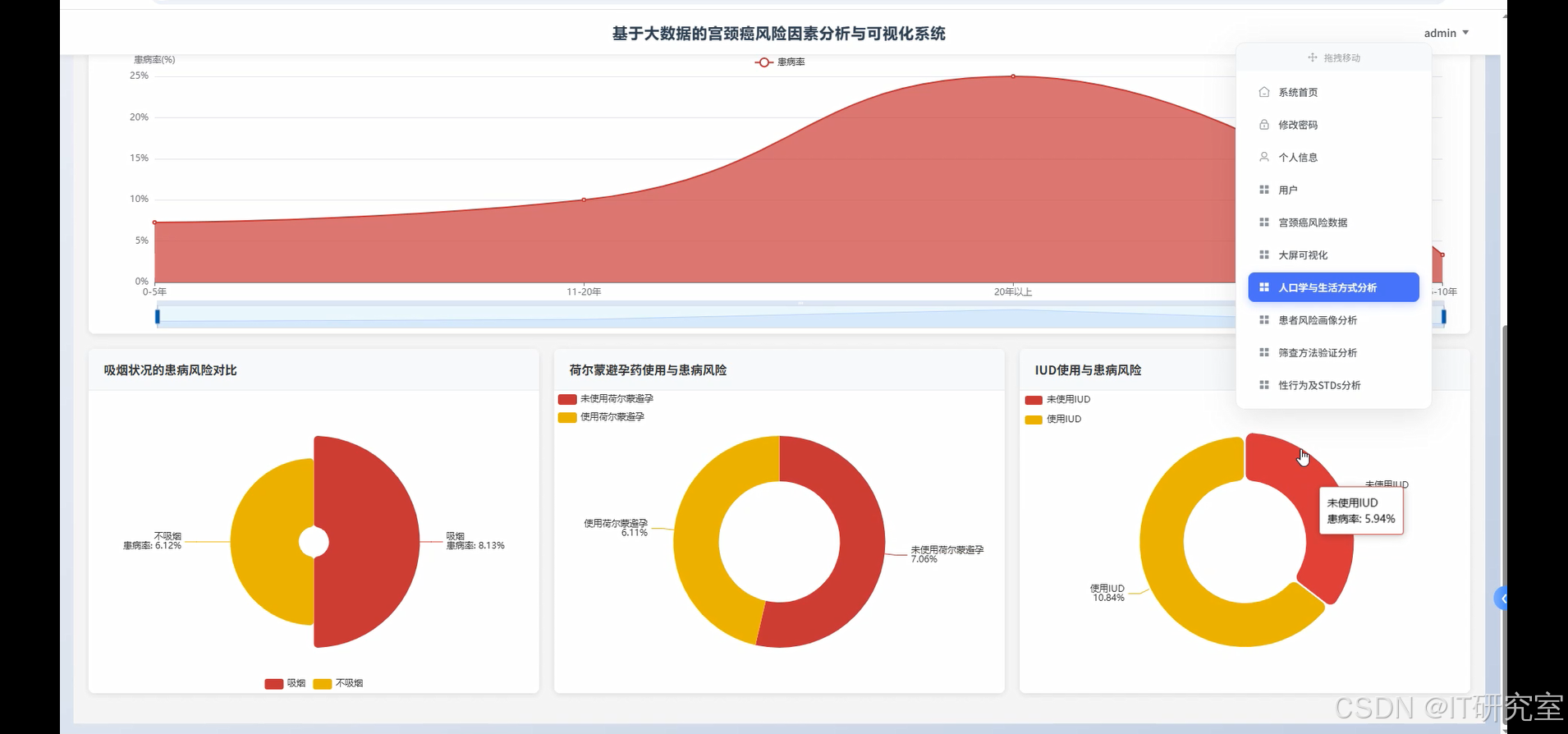
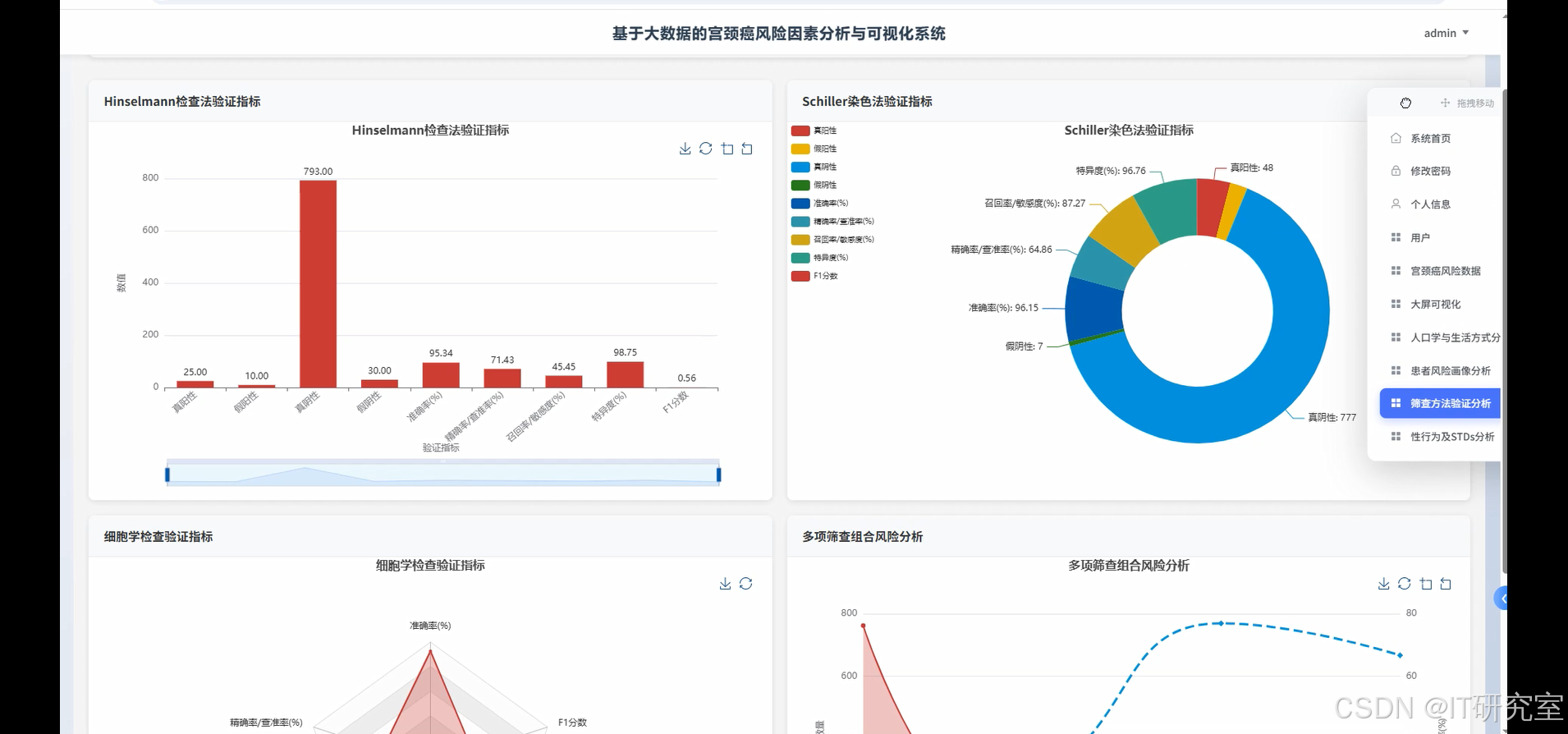
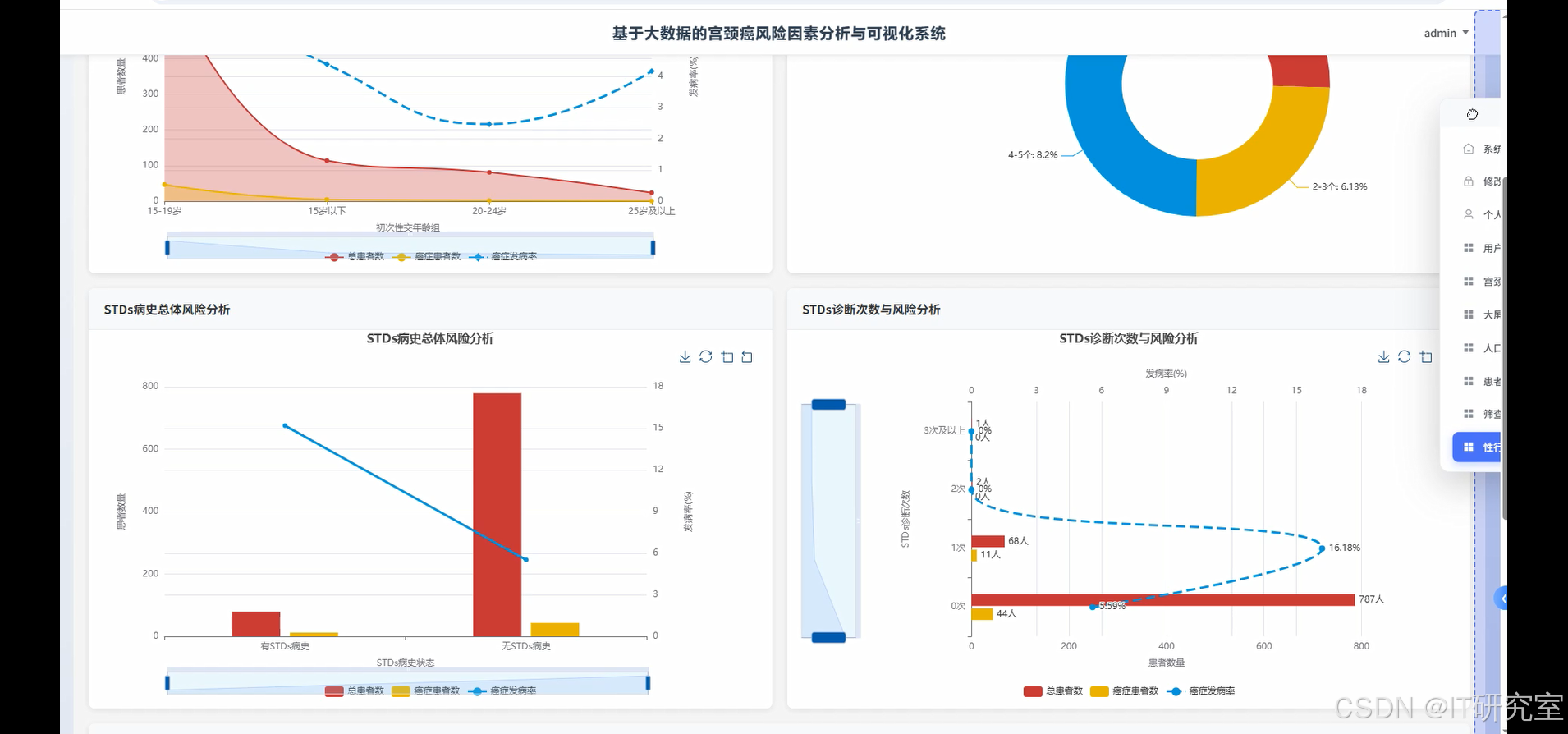
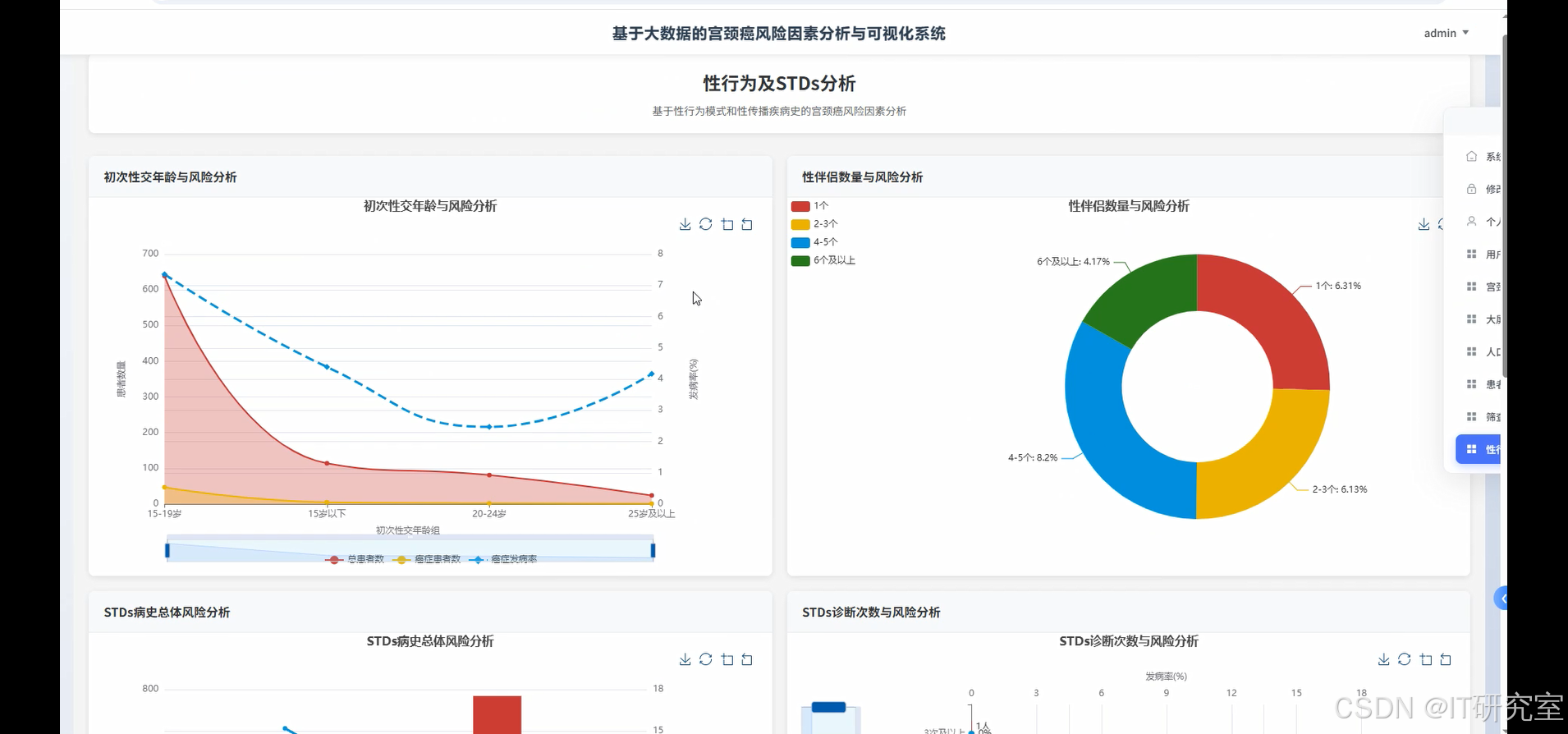
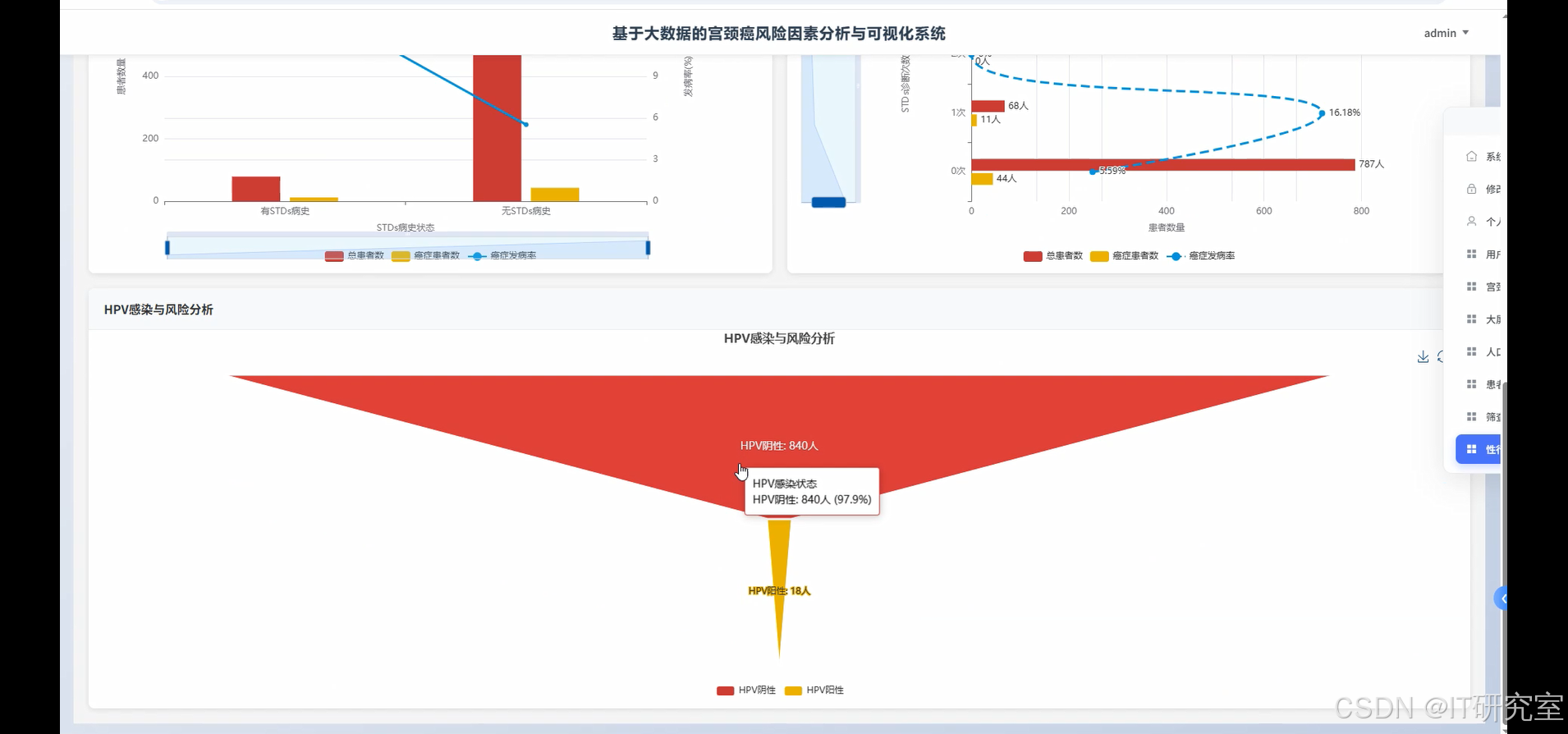
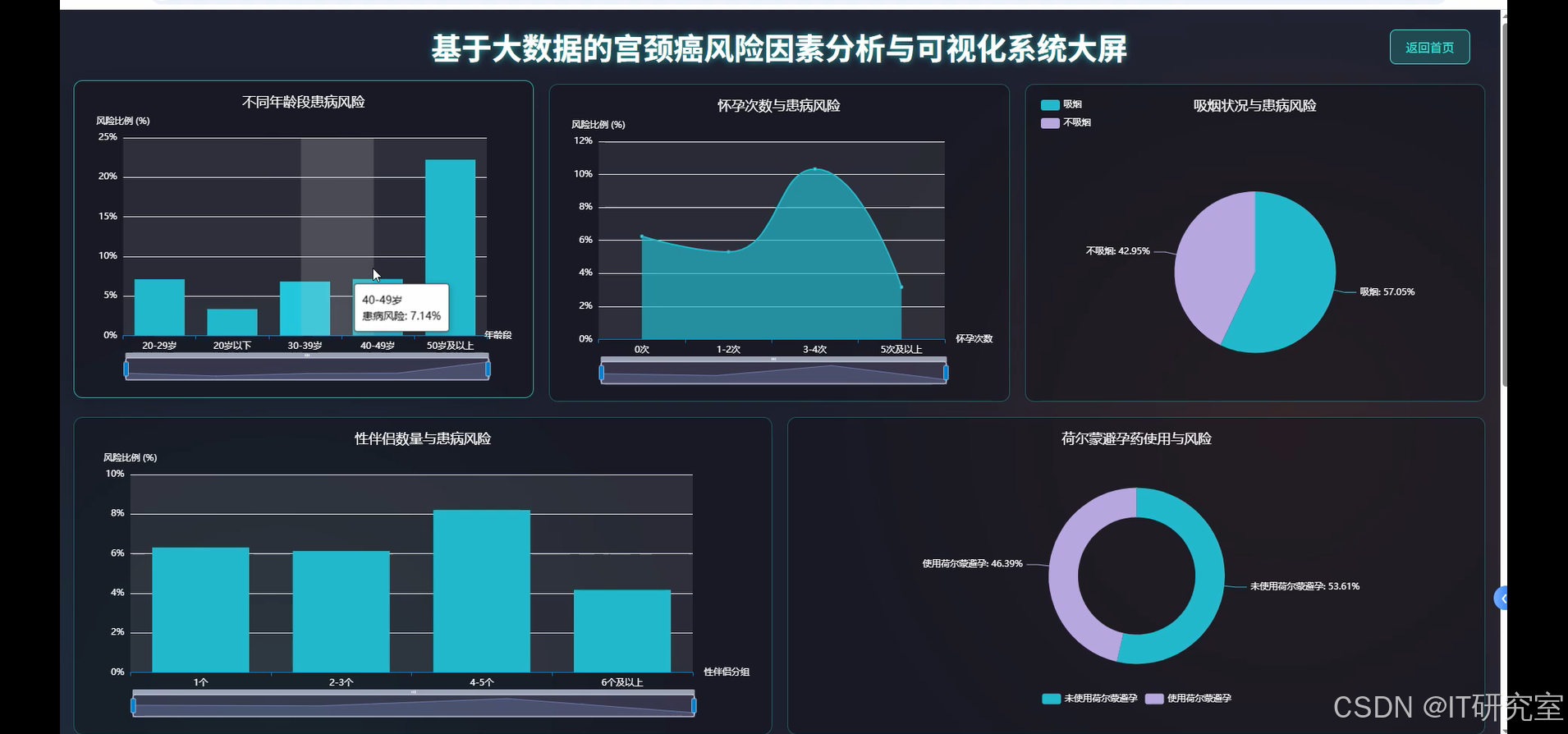
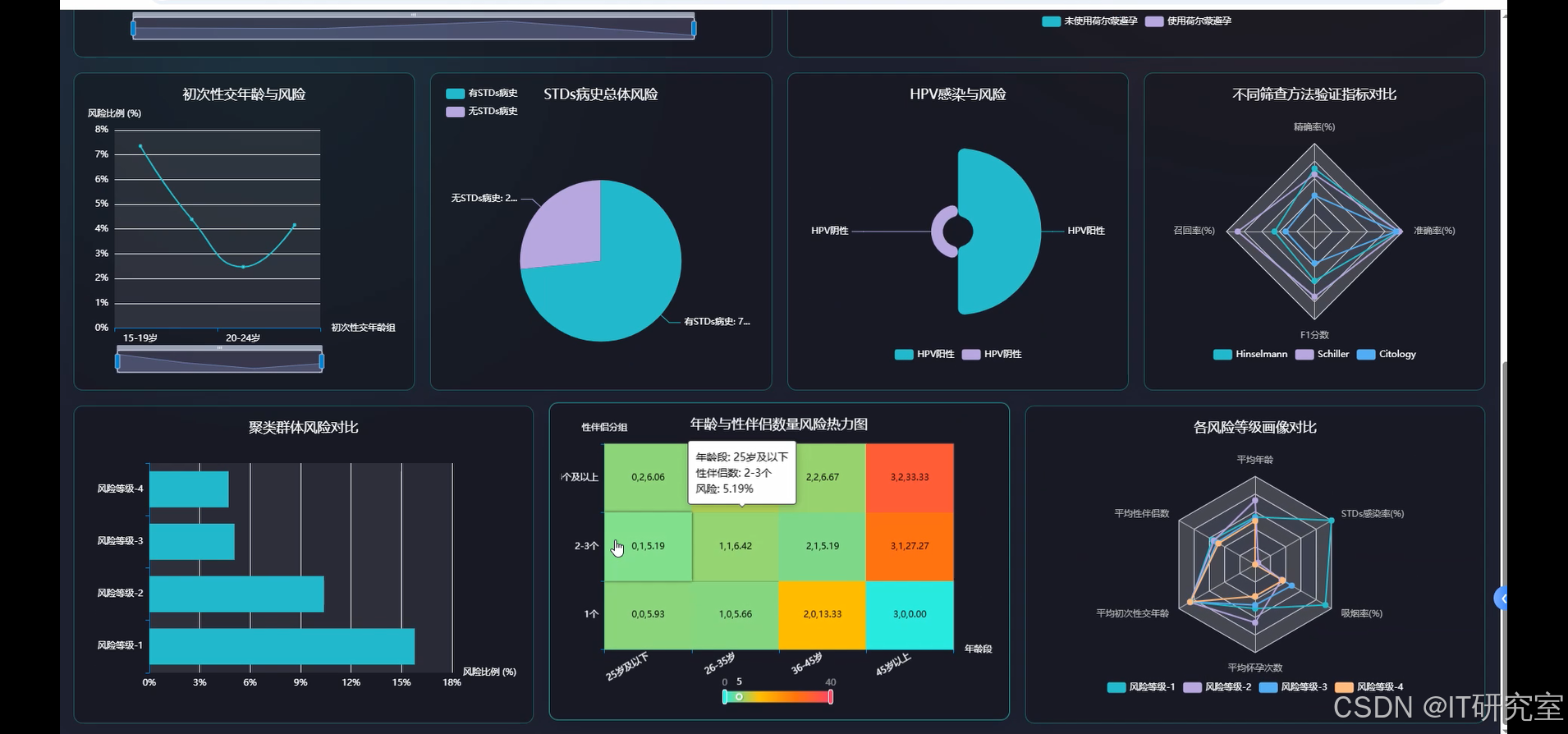
四、代码参考
- 项目实战代码参考:
java(贴上部分代码)
from pyspark.sql import SparkSession
from pyspark.ml.clustering import KMeans
from pyspark.ml.feature import VectorAssembler
from pyspark.sql.functions import col, count, avg, when, desc, asc
import pandas as pd
import numpy as np
from django.http import JsonResponse
from django.views import View
import json
spark = SparkSession.builder.appName("CervicalCancerRiskAnalysis").config("spark.sql.adaptive.enabled", "true").config("spark.sql.adaptive.coalescePartitions.enabled", "true").getOrCreate()
class PatientRiskProfileAnalysis(View):
def post(self, request):
data = json.loads(request.body)
df = spark.read.csv("hdfs://localhost:9000/cervical_cancer_data/risk_factors_cervical_cancer.csv", header=True, inferSchema=True)
df_cleaned = df.filter(col("Age").isNotNull() & col("Number of sexual partners").isNotNull() & col("Num of pregnancies").isNotNull())
feature_cols = ["Age", "Number of sexual partners", "First sexual intercourse", "Num of pregnancies", "Smokes", "STDs"]
assembler = VectorAssembler(inputCols=feature_cols, outputCol="features")
df_features = assembler.transform(df_cleaned.fillna(0))
kmeans = KMeans(k=4, seed=42, featuresCol="features", predictionCol="risk_cluster")
model = kmeans.fit(df_features)
clustered_df = model.transform(df_features)
cluster_stats = clustered_df.groupBy("risk_cluster").agg(
count("*").alias("patient_count"),
avg("Age").alias("avg_age"),
avg("Number of sexual partners").alias("avg_partners"),
avg("Biopsy").alias("cancer_rate")
).orderBy("cancer_rate", ascending=False)
age_distribution = clustered_df.groupBy("risk_cluster").agg(
count(when(col("Age") < 25, 1)).alias("age_under_25"),
count(when((col("Age") >= 25) & (col("Age") < 35), 1)).alias("age_25_35"),
count(when((col("Age") >= 35) & (col("Age") < 45), 1)).alias("age_35_45"),
count(when(col("Age") >= 45, 1)).alias("age_over_45")
)
risk_indicators = clustered_df.groupBy("risk_cluster").agg(
avg("STDs").alias("avg_stds_rate"),
avg("Smokes").alias("avg_smoking_rate"),
avg("Hormonal Contraceptives").alias("avg_contraceptive_use")
)
cluster_summary = cluster_stats.join(age_distribution, "risk_cluster").join(risk_indicators, "risk_cluster")
result_data = cluster_summary.collect()
response_data = []
for row in result_data:
cluster_info = {
"cluster_id": row["risk_cluster"],
"patient_count": row["patient_count"],
"cancer_rate": round(row["cancer_rate"] * 100, 2),
"avg_age": round(row["avg_age"], 1),
"avg_partners": round(row["avg_partners"], 1),
"age_distribution": {
"under_25": row["age_under_25"],
"25_35": row["age_25_35"],
"35_45": row["age_35_45"],
"over_45": row["age_over_45"]
},
"risk_indicators": {
"stds_rate": round(row["avg_stds_rate"] * 100, 2),
"smoking_rate": round(row["avg_smoking_rate"] * 100, 2),
"contraceptive_rate": round(row["avg_contraceptive_use"] * 100, 2)
}
}
response_data.append(cluster_info)
return JsonResponse({"clusters": response_data, "total_patients": df_cleaned.count()})
class ScreeningMethodValidation(View):
def post(self, request):
data = json.loads(request.body)
df = spark.read.csv("hdfs://localhost:9000/cervical_cancer_data/risk_factors_cervical_cancer.csv", header=True, inferSchema=True)
df_screening = df.select("Hinselmann", "Schiller", "Citology", "Biopsy").filter(
col("Hinselmann").isNotNull() & col("Schiller").isNotNull() &
col("Citology").isNotNull() & col("Biopsy").isNotNull()
)
methods = ["Hinselmann", "Schiller", "Citology"]
validation_results = {}
for method in methods:
tp = df_screening.filter((col(method) == 1) & (col("Biopsy") == 1)).count()
tn = df_screening.filter((col(method) == 0) & (col("Biopsy") == 0)).count()
fp = df_screening.filter((col(method) == 1) & (col("Biopsy") == 0)).count()
fn = df_screening.filter((col(method) == 0) & (col("Biopsy") == 1)).count()
total = tp + tn + fp + fn
accuracy = (tp + tn) / total if total > 0 else 0
sensitivity = tp / (tp + fn) if (tp + fn) > 0 else 0
specificity = tn / (tn + fp) if (tn + fp) > 0 else 0
precision = tp / (tp + fp) if (tp + fp) > 0 else 0
f1_score = 2 * (precision * sensitivity) / (precision + sensitivity) if (precision + sensitivity) > 0 else 0
validation_results[method] = {
"accuracy": round(accuracy * 100, 2),
"sensitivity": round(sensitivity * 100, 2),
"specificity": round(specificity * 100, 2),
"precision": round(precision * 100, 2),
"f1_score": round(f1_score, 3),
"confusion_matrix": {
"true_positive": tp,
"true_negative": tn,
"false_positive": fp,
"false_negative": fn
}
}
combined_positive = df_screening.filter(
(col("Hinselmann") == 1) | (col("Schiller") == 1) | (col("Citology") == 1)
)
combined_analysis = combined_positive.groupBy("Biopsy").count().collect()
positive_combinations = df_screening.withColumn(
"positive_methods_count",
col("Hinselmann") + col("Schiller") + col("Citology")
).groupBy("positive_methods_count", "Biopsy").count().collect()
combination_analysis = {}
for row in positive_combinations:
method_count = row["positive_methods_count"]
if method_count not in combination_analysis:
combination_analysis[method_count] = {"positive_biopsy": 0, "negative_biopsy": 0}
if row["Biopsy"] == 1:
combination_analysis[method_count]["positive_biopsy"] = row["count"]
else:
combination_analysis[method_count]["negative_biopsy"] = row["count"]
return JsonResponse({
"individual_methods": validation_results,
"combination_analysis": combination_analysis,
"total_samples": df_screening.count()
})
class DemographicLifestyleAnalysis(View):
def post(self, request):
data = json.loads(request.body)
df = spark.read.csv("hdfs://localhost:9000/cervical_cancer_data/risk_factors_cervical_cancer.csv", header=True, inferSchema=True)
df_clean = df.filter(col("Age").isNotNull() & col("Biopsy").isNotNull())
age_groups = df_clean.withColumn(
"age_group",
when(col("Age") < 20, "Under 20")
.when((col("Age") >= 20) & (col("Age") < 30), "20-29")
.when((col("Age") >= 30) & (col("Age") < 40), "30-39")
.when((col("Age") >= 40) & (col("Age") < 50), "40-49")
.otherwise("50+")
)
age_analysis = age_groups.groupBy("age_group").agg(
count("*").alias("total_patients"),
count(when(col("Biopsy") == 1, 1)).alias("positive_cases"),
avg("Biopsy").alias("cancer_rate")
).orderBy("age_group")
pregnancy_analysis = df_clean.filter(col("Num of pregnancies").isNotNull()).withColumn(
"pregnancy_group",
when(col("Num of pregnancies") == 0, "0")
.when((col("Num of pregnancies") >= 1) & (col("Num of pregnancies") <= 2), "1-2")
.when((col("Num of pregnancies") >= 3) & (col("Num of pregnancies") <= 5), "3-5")
.otherwise("6+")
).groupBy("pregnancy_group").agg(
count("*").alias("total_patients"),
avg("Biopsy").alias("cancer_rate")
).orderBy("pregnancy_group")
smoking_analysis = df_clean.filter(col("Smokes").isNotNull()).groupBy("Smokes").agg(
count("*").alias("total_patients"),
avg("Biopsy").alias("cancer_rate")
).collect()
smoking_detailed = df_clean.filter((col("Smokes") == 1) & col("Smokes (years)").isNotNull()).withColumn(
"smoking_years_group",
when(col("Smokes (years)") < 5, "Less than 5 years")
.when((col("Smokes (years)") >= 5) & (col("Smokes (years)") < 15), "5-15 years")
.when((col("Smokes (years)") >= 15) & (col("Smokes (years)") < 25), "15-25 years")
.otherwise("25+ years")
).groupBy("smoking_years_group").agg(
count("*").alias("total_patients"),
avg("Biopsy").alias("cancer_rate"),
avg("Smokes (packs/year)").alias("avg_packs_per_year")
).collect()
contraceptive_analysis = df_clean.filter(col("Hormonal Contraceptives").isNotNull()).groupBy("Hormonal Contraceptives").agg(
count("*").alias("total_patients"),
avg("Biopsy").alias("cancer_rate")
).collect()
iud_analysis = df_clean.filter(col("IUD").isNotNull()).groupBy("IUD").agg(
count("*").alias("total_patients"),
avg("Biopsy").alias("cancer_rate")
).collect()
age_data = [{"age_group": row["age_group"], "total_patients": row["total_patients"], "positive_cases": row["positive_cases"], "cancer_rate": round(row["cancer_rate"] * 100, 2)} for row in age_analysis.collect()]
pregnancy_data = [{"pregnancy_group": row["pregnancy_group"], "total_patients": row["total_patients"], "cancer_rate": round(row["cancer_rate"] * 100, 2)} for row in pregnancy_analysis.collect()]
smoking_data = [{"smoking_status": "Smoker" if row["Smokes"] == 1 else "Non-smoker", "total_patients": row["total_patients"], "cancer_rate": round(row["cancer_rate"] * 100, 2)} for row in smoking_analysis]
smoking_years_data = [{"years_group": row["smoking_years_group"], "total_patients": row["total_patients"], "cancer_rate": round(row["cancer_rate"] * 100, 2), "avg_packs_per_year": round(row["avg_packs_per_year"], 2)} for row in smoking_detailed]
contraceptive_data = [{"method": "Hormonal" if row["Hormonal Contraceptives"] == 1 else "None", "total_patients": row["total_patients"], "cancer_rate": round(row["cancer_rate"] * 100, 2)} for row in contraceptive_analysis]
iud_data = [{"method": "IUD" if row["IUD"] == 1 else "No IUD", "total_patients": row["total_patients"], "cancer_rate": round(row["cancer_rate"] * 100, 2)} for row in iud_analysis]
return JsonResponse({
"age_analysis": age_data,
"pregnancy_analysis": pregnancy_data,
"smoking_analysis": smoking_data,
"smoking_years_analysis": smoking_years_data,
"contraceptive_analysis": contraceptive_data,
"iud_analysis": iud_data,
"total_patients": df_clean.count()
})五、系统视频
基于大数据的宫颈癌风险因素分析与可视化系统项目视频:
大数据毕业设计选题推荐-基于大数据的宫颈癌风险因素分析与可视化系统-Spark-Hadoop-Bigdata
结语
大数据毕业设计选题推荐-基于大数据的宫颈癌风险因素分析与可视化系统-Spark-Hadoop-Bigdata
想看其他类型的计算机毕业设计作品也可以和我说~谢谢大家!
有技术这一块问题大家可以评论区交流或者私我~
大家可以帮忙点赞、收藏、关注、评论啦~
源码获取:⬇⬇⬇