文章目录
- 先放结论
- 背景
- Pubmed数据库的访问
- 一,Bio.Entrez
-
- 1,借助LibInspector
- 2,回到BioPython文档
-
- [1,EUtils XML结果的4种解析方法](#1,EUtils XML结果的4种解析方法)
- [2,NCBI DTD文件的作用与缺失/过时处理](#2,NCBI DTD文件的作用与缺失/过时处理)
-
- [1. DTD文件的核心作用](#1. DTD文件的核心作用)
- [2. DTD文件的常见问题与解决方案](#2. DTD文件的常见问题与解决方案)
- [3,通过Biopython访问NCBI的线上资源规范(NCBI的Entrez 用户规范)](#3,通过Biopython访问NCBI的线上资源规范(NCBI的Entrez 用户规范))
- [4,EInfo: 获取Entrez数据库的信息](#4,EInfo: 获取Entrez数据库的信息)
-
- [1. einfo.dtd:定义XML的结构规则](#1. einfo.dtd:定义XML的结构规则)
- [2. XML示例:eInfo的实际返回结果](#2. XML示例:eInfo的实际返回结果)
- [3. 实际用途:如何用这些信息?](#3. 实际用途:如何用这些信息?)
-
- [1. 开发层面:解析eInfo返回数据](#1. 开发层面:解析eInfo返回数据)
- [2. 使用层面:了解数据库检索规则](#2. 使用层面:了解数据库检索规则)
- [4. 将这个XML读入到一个Python对象里面去](#4. 将这个XML读入到一个Python对象里面去)
- [5,ESearch: 搜索Entrez数据库](#5,ESearch: 搜索Entrez数据库)
- [6,EPost: 上传identifiers的列表](#6,EPost: 上传identifiers的列表)
- [7,ESummary: 通过主要的IDs来获取摘要](#7,ESummary: 通过主要的IDs来获取摘要)
- [8,EFetch: 从Entrez下载更多的记录](#8,EFetch: 从Entrez下载更多的记录)
- [9,ELink: 在NCBI Entrez中搜索相关的条目](#9,ELink: 在NCBI Entrez中搜索相关的条目)
- [10,EGQuery: 全局搜索- 统计搜索的条目](#10,EGQuery: 全局搜索- 统计搜索的条目)
- [11,ESpell: 获得拼写建议](#11,ESpell: 获得拼写建议)
- [二,Entrez Direct](#二,Entrez Direct)
先放结论
这篇博客我打算出成一个系列,就讲如何仔细拆分文献阅读任务,如何将文献阅读流程化、自动化。
我觉得,在AI时代,这个科研人员的小终极目标,是已经有一些初步的解决办法的。
背景
前面出了几篇博客讲如何找代码、如何自动化阅读/解析代码,
这些都是下游的任务,作为科研人员,除了输出+执行之外,不可避免的还得大量阅读文献输入。
那么,文献阅读可不可以自动化呢,可不可以转换为一种形而下的纯体力劳动呢?
答案是:可以的。
所谓体力劳动化,其实我们第一个想到的就是编程,就是如何让任务流程自动化,只要能够转换为计算机流程处理的任务,那么阅读文献其实就从一个费时费力、形而上又脑力劳动的任务变成了一个体力劳动,这一点,我想对于大部分科研人员来说大家都是很乐意见到的。
所以我就在想,如何将文献阅读,变成一个计算可执行的任务?
或者,现实一点地讲,如何理清我们文献阅读的流程,并依据这个流程将需求进行自动化、智能化处理。
下面是我的一个初步的想法,其中的阶段是我个人认为的文献阅读的一个完整生命周期链,也就是我理想中的文献阅读的流程化,只有将文献阅读这个任务给本体论化了,我们才能抽象出具体的编程需求。仅代表个人意见,仅供参考。
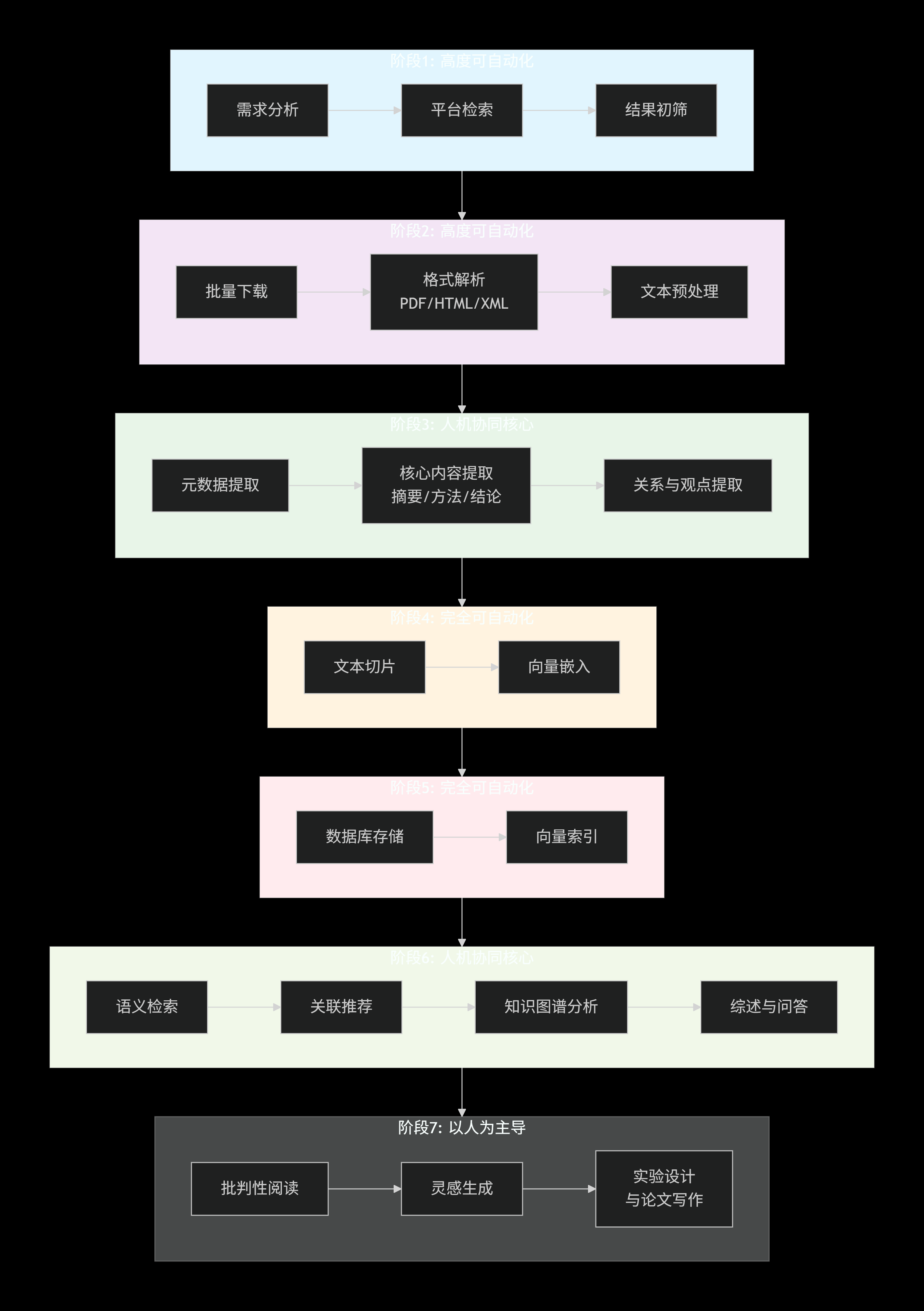
因为重点在于自动化,只不过这整个流程的遍历需要借助AI/LLM,所以不一定每一节都会加AI。
本篇博客先来讲一讲文献检索如何自动化,我们以生物医学领域(AI4S需求最大的群体之一)的pubmed文献数据库为例进行展开。
Pubmed数据库的访问
对于Pubmed数据库的访问,我们先来讲一下如何访问访问NCBI Entrez数据库。
因为Pubmed本质上也只是一个NCBI系的数据库,而Entrez则是NCBI官方通用的检索系统。
Entrez ( http://www.ncbi.nlm.nih.gov/Entrez)) 是一个给客户提供NCBI各个数据库(如PubMed, GeneBank, GEO等等)访问的检索系统。 用户可以通过浏览器手动输入查询条目访问Entrez,也可以使用Biopython的 Bio.Entrez 模块以编程方式访问来访问Entrez。 如果使用第二种方法,用户用一个Python脚本就可以实现在PubMed里面搜索或者从GenBank下载数据。
Bio.Entrez 模块利用了Entrez Programming Utilities(也称作EUtils),包含八个工具,详情请见NCBI的网站: http://www.ncbi.nlm.nih.gov/entrez/utils/. 每个工具都能在Python的 Bio.Entrez 模块中找到对应函数,后面会详细讲到。这个模块可以保证用来查询的URL 的正确性,并且向NCBI要求的一样,每三秒钟查询的次数不超过一。
简而言之,BioPython中的Entrez模块是对Entrez Direct的套壳,
熟悉python的可以直接投入BioPython,或者用python自己的XML解析模块解析Entrez Direct返回的内容;
熟悉Awk之类shell脚本语言的,可以直接上手Entrez Direct,其本身也有一套类似体系的脚本语言形式。
一,Bio.Entrez
很显然,我们编程的重点在于Bio.Entrez,
我们可以先看一下这个模块的一些细节
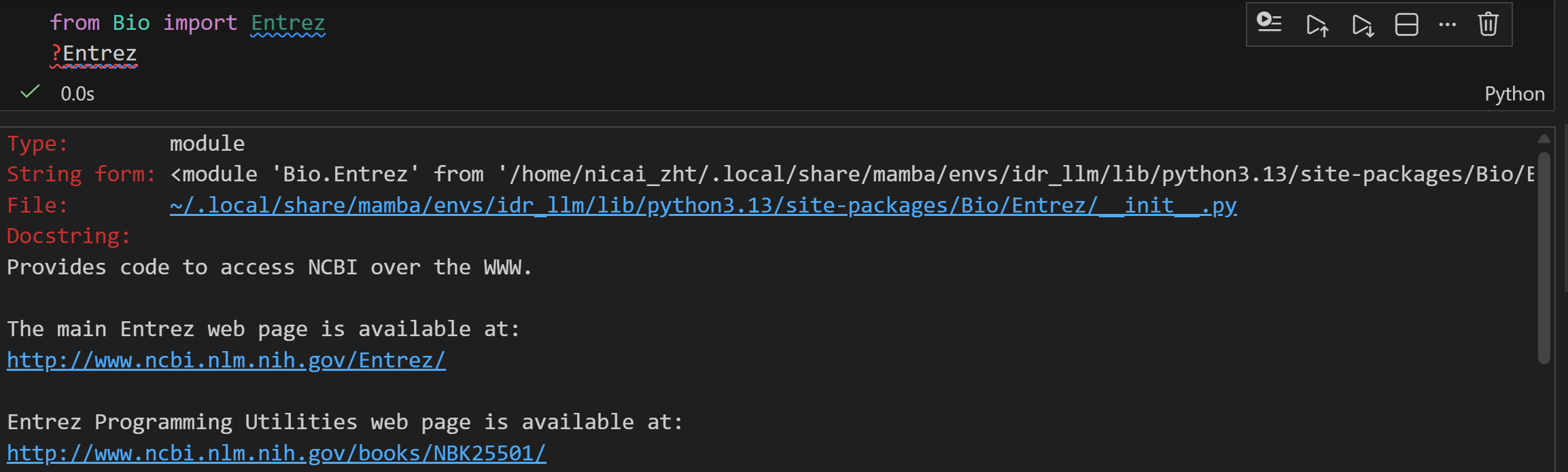


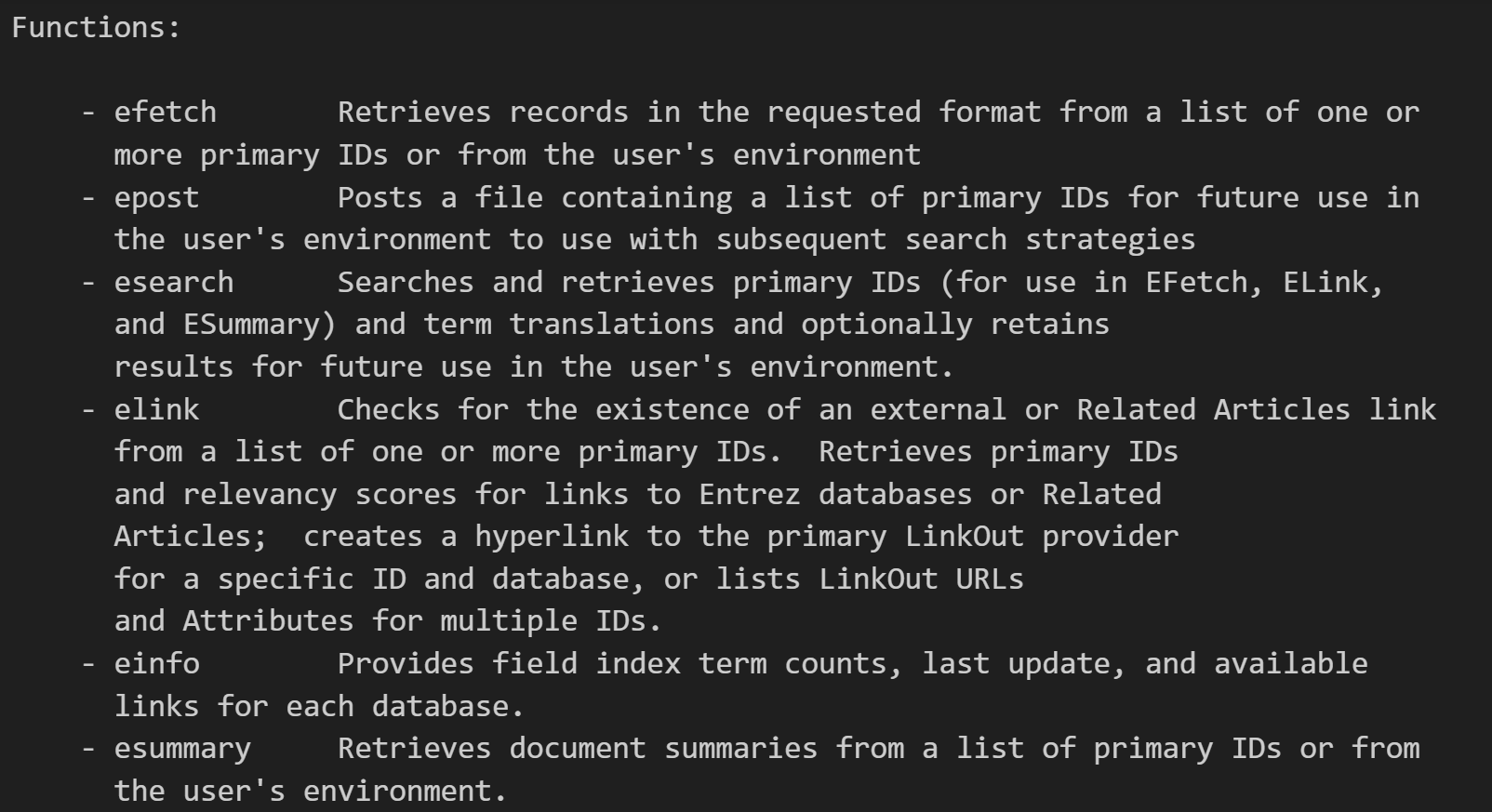
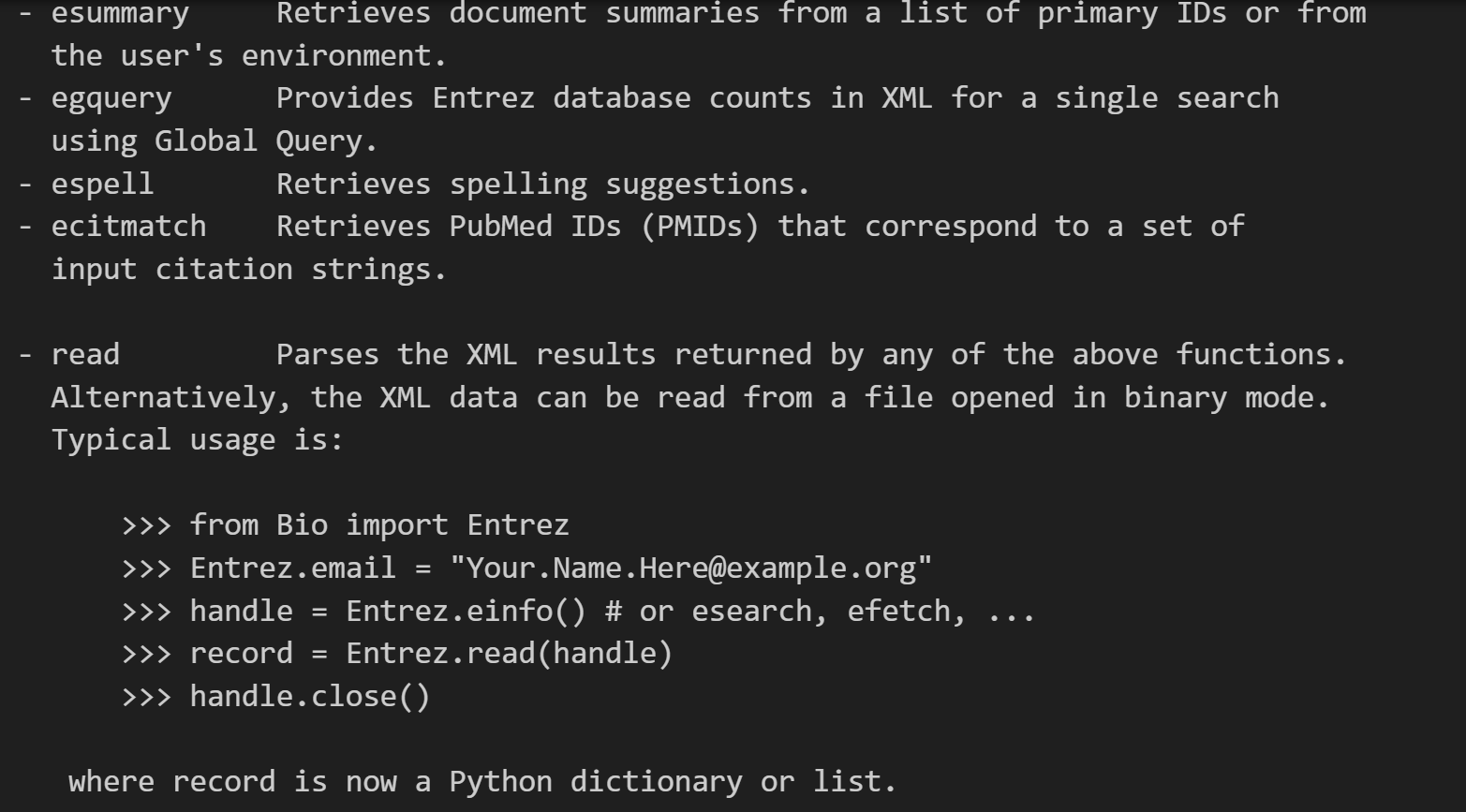
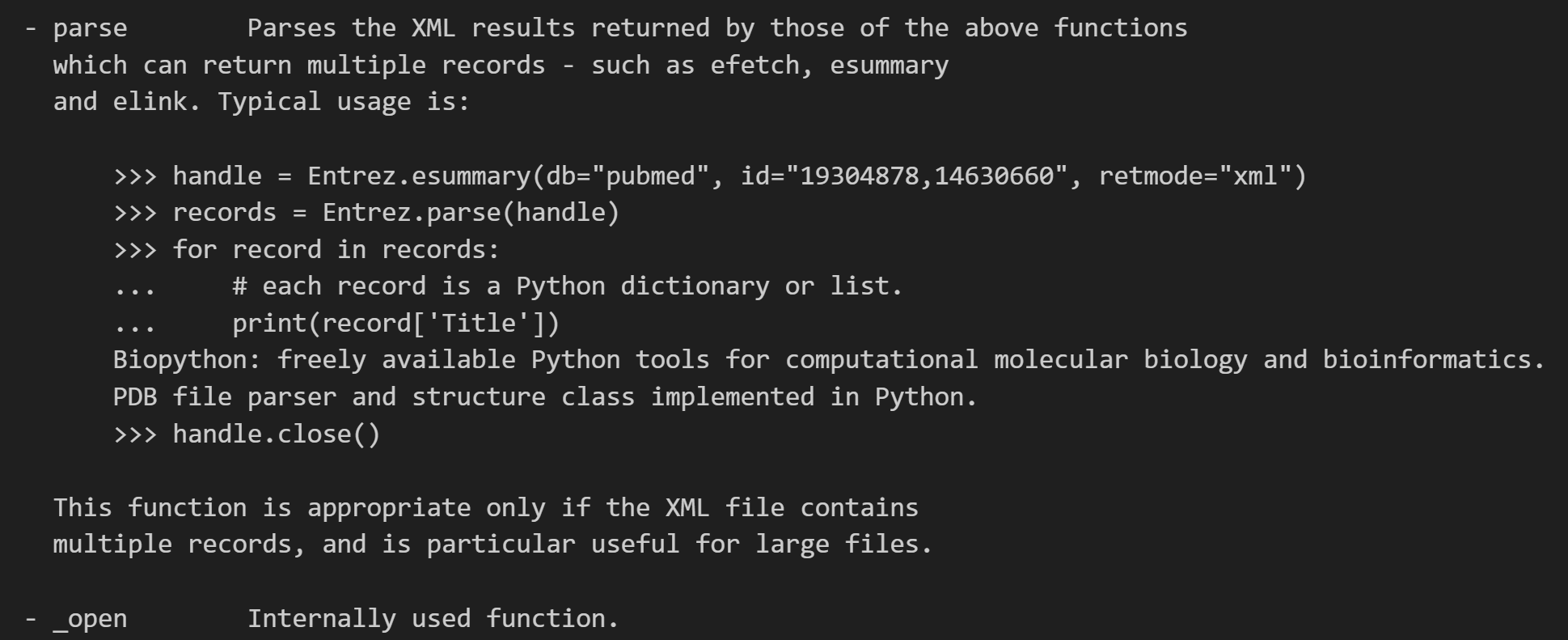
其实从这个文档解释中我们就已经能够看到很多了,
包括一些最基础的用法:
最好还是设置一下我们的email
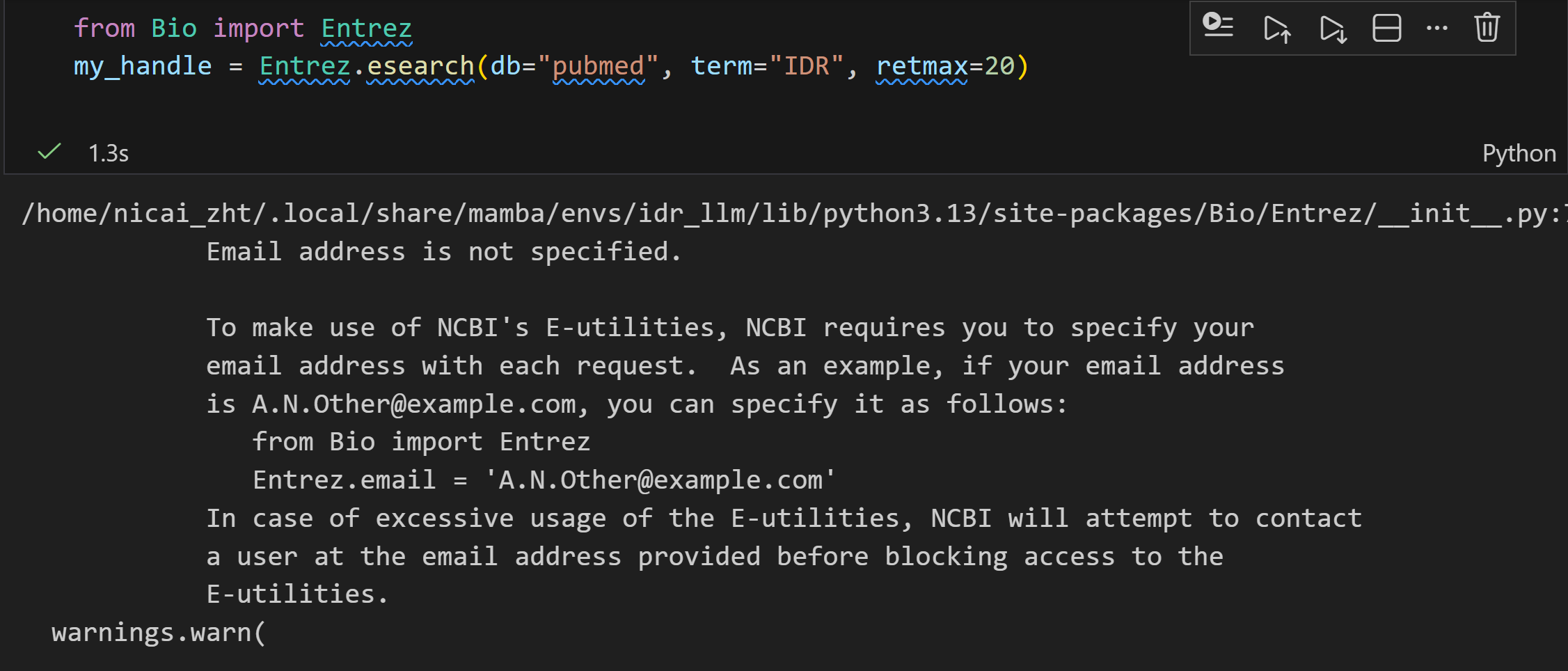
加上邮箱之后返回的
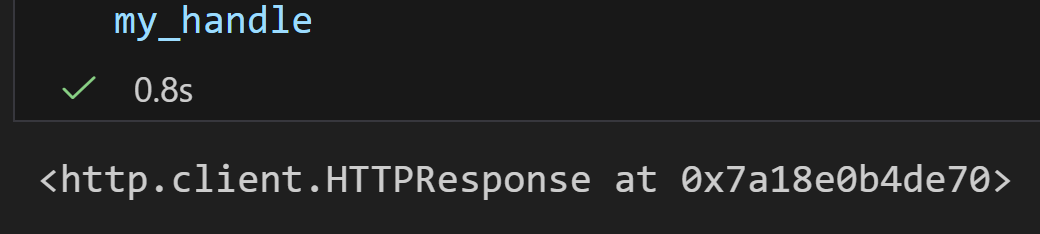
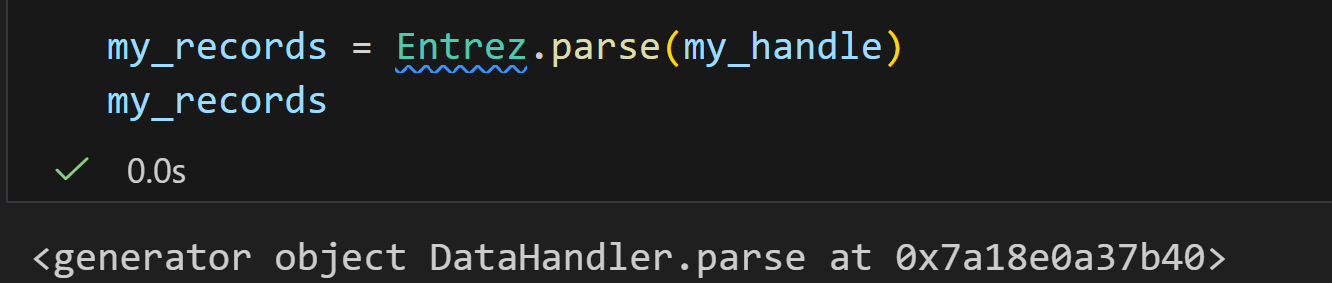
但是报错:
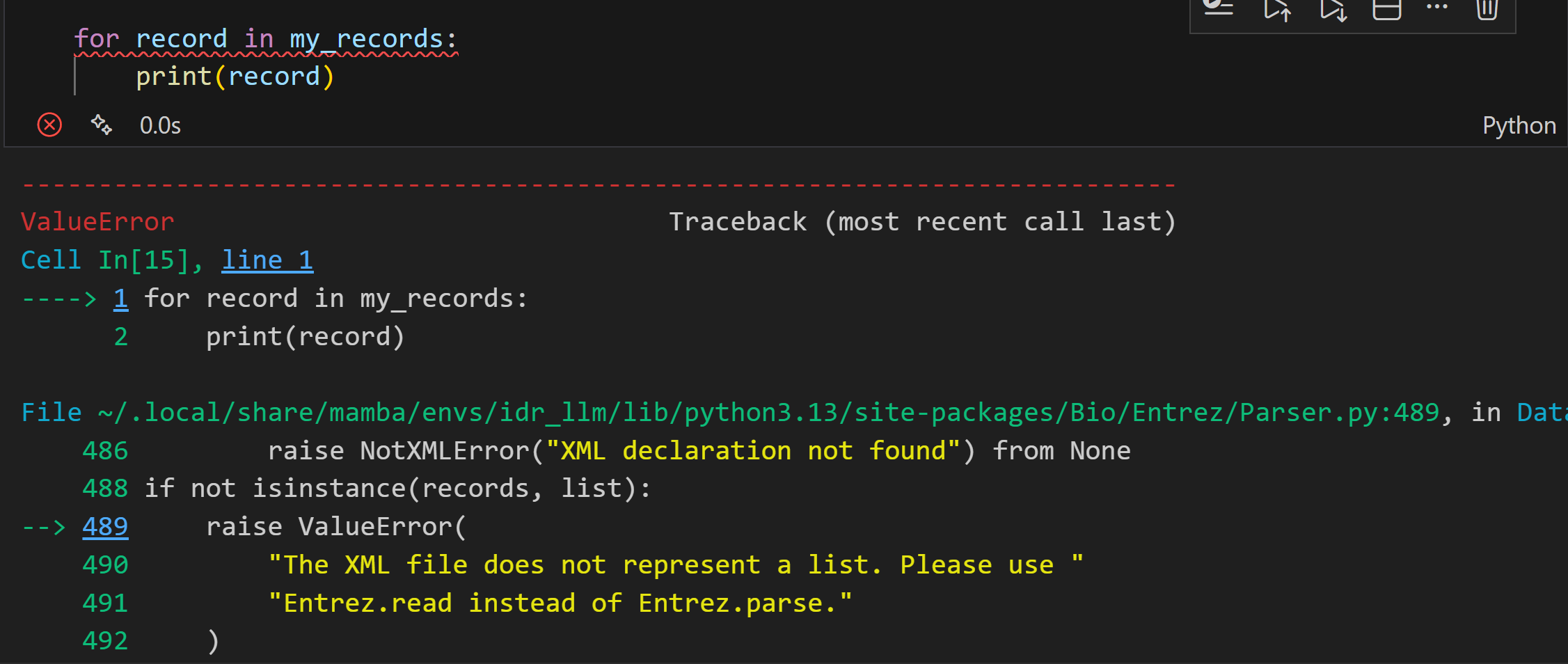

后来检查才发现:
这个错误是因为 Entrez.esearch 返回的是单个搜索结果(一个包含 ID 和元数据的 XML 结构),而不是多个记录的列表。Entrez.parse 设计用于处理多个记录的生成器(例如来自 efetch 的结果),而 Entrez.read 用于处理单个 XML 结果。
所有逻辑就是:我是用esearch返回单个搜索结果,再用read处理这个单个的XML结果。
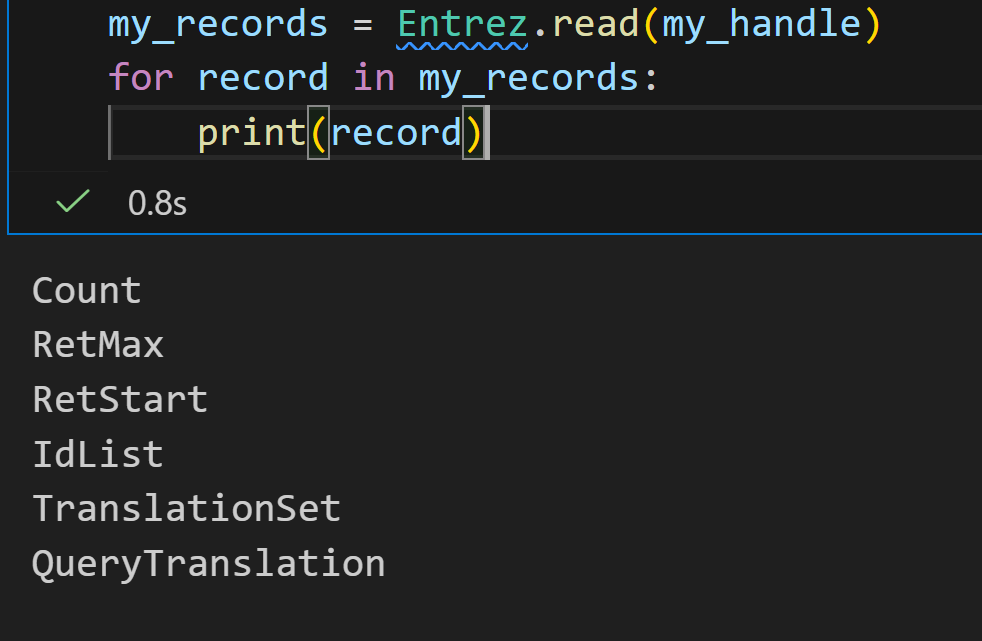
将每一个键值对打印出来,发现结果晦涩难懂的:
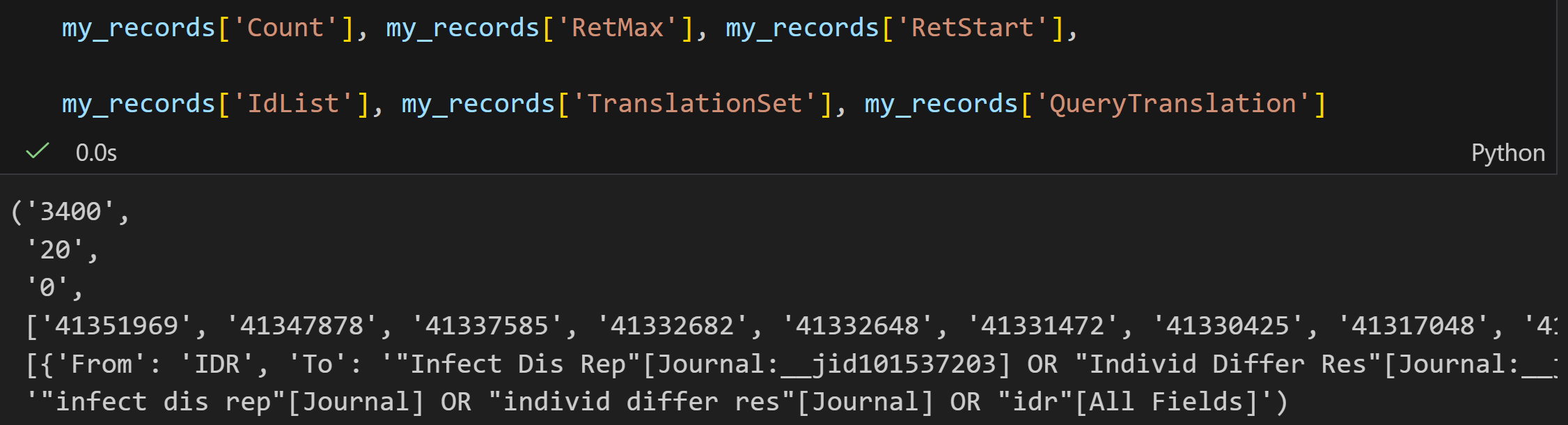
1,借助LibInspector
此处,我们可以先查看一下Bio.Entrez这个模块一下,
使用我之前开发的工具,具体可以查看我之前的博客:LibInspector---为小白智能解析、阅读(几乎所有)Python工具库,细节文档说明可以查看该工具的开发仓库
bash
❯ lib-inspector Bio.Entrez -o ./test/entrz.md
🔍 Target Resolution: Input='Bio.Entrez' -> Import='Bio.Entrez' (Type: module)
🔍 Analyzing dependencies for 'Bio.Entrez' (Network Analysis Phase)...
✅ Markdown report saved to: /data2/LibInspector/test/entrz.md
📊 Interactive HTML report saved to: /data2/LibInspector/test/entrz.html
(Open the HTML file in your browser to see rendered charts)结果的md以及html文件我已经放在仓库https://github.com/MaybeBio/LibInspector/tree/main/test** **里了;
我们直接看输出报告文件的Navigator:
基本上把一些重要的API都列出来了,当然,我这里限制20个:
这些API我们可以一一再?去检索具体的逻辑
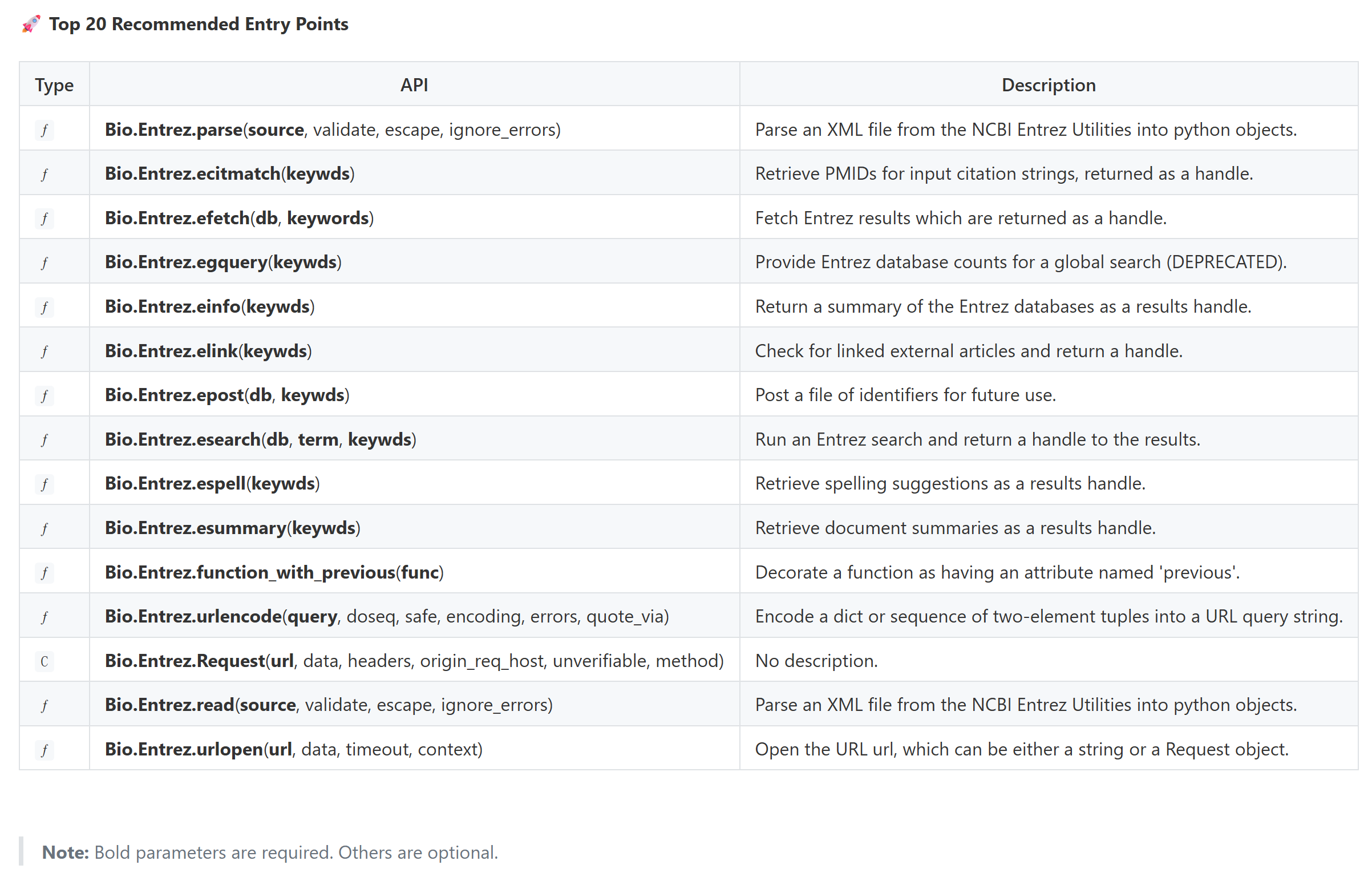
当然,具体有用的api其实我工具设计的时候就考虑到了,可以直接在top-level api部分获取:
我再补充一下,设计top-level这一块的目的,工具生成「库 API 清单」的基础 ------ 通过筛选排除无效 / 冗余成员,再按类型分类,后续可基于 classes 和 functions 生成结构化文档(如展示类的方法、函数的签名和文档字符串),帮助用户快速了解库的核心功能。
具体有两个参数限制,大家可以自行按照我文档中的提示去设置,

总之大概API解析的结果我输出文档中已经有了,如下:
ecitmatch
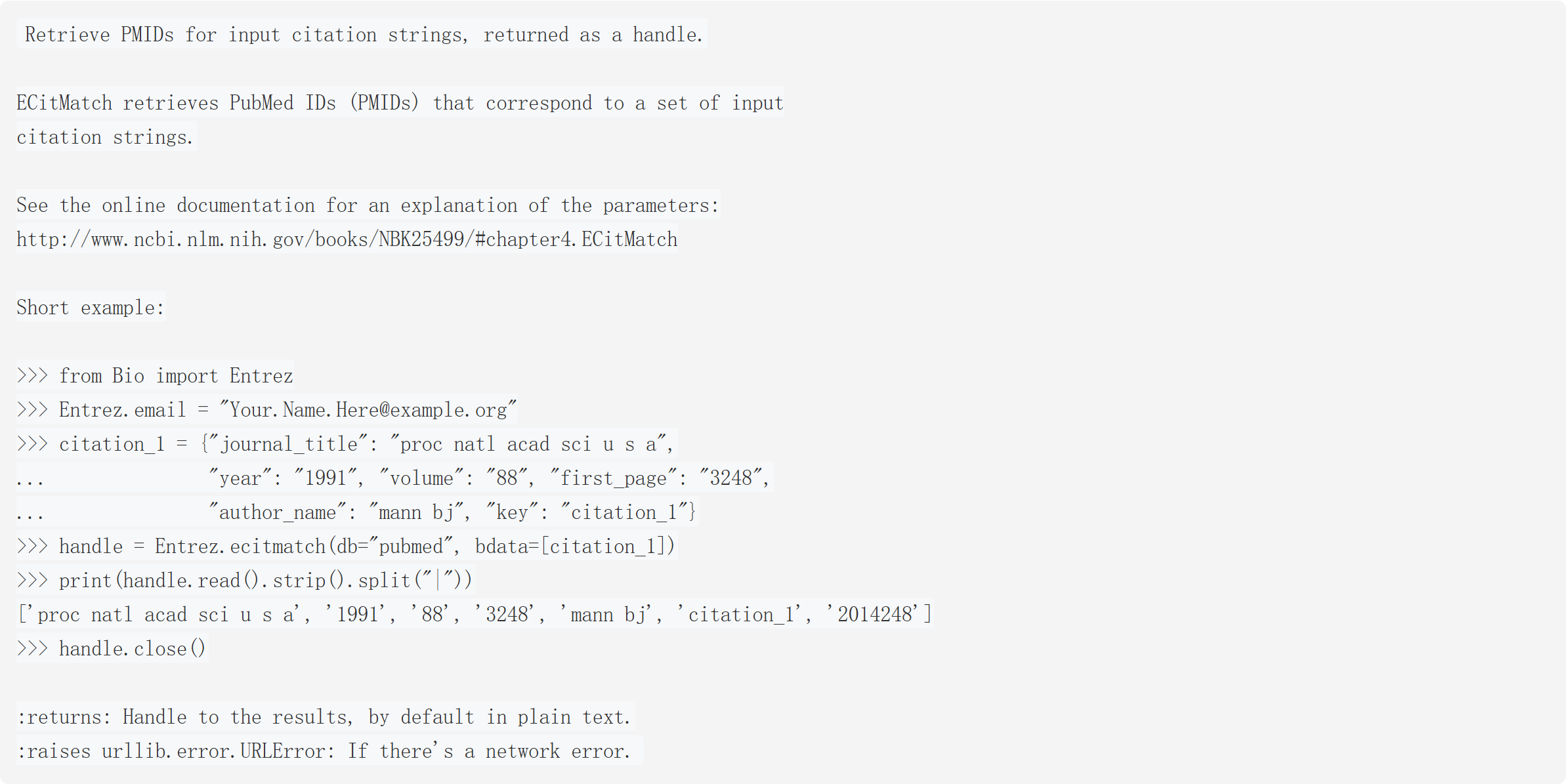
这段代码 / 文档的核心是介绍 NCBI Entrez 库的**ecitmatch**函数:
它的作用是根据输入的文献引用信息(期刊名、年份、卷、起始页、作者名等),检索对应的 PubMed ID (PMID)(PMID 是 PubMed 数据库给每篇文献分配的唯一标识),最终返回一个可读取结果的 "句柄(handle)"。
bash
from Bio import Entrez # 导入Biopython的Entrez模块(用于访问NCBI数据库)
Entrez.email = "Your.Name.Here@example.org" # 必须填写邮箱(NCBI要求,用于追踪访问)
# 定义一条引用信息的字典
citation_1 = {
"journal_title": "proc natl acad sci u s a", # 期刊名(需小写,缩写/全称均可)
"year": "1991", # 发表年份
"volume": "88", # 卷号
"first_page": "3248", # 起始页码
"author_name": "mann bj", # 作者名(姓+名首字母,小写)
"key": "citation_1" # 自定义标识(用于匹配结果和输入)
}
# 调用ecitmatch函数,请求PMID匹配
handle = Entrez.ecitmatch(db="pubmed", bdata=[citation_1])
# 读取结果并处理:按|分割字符串
print(handle.read().strip().split("|"))
# 输出:['proc natl acad sci u s a', '1991', '88', '3248', 'mann bj', 'citation_1', '2014248']
# 最后一个元素"2014248"就是匹配到的PMID
handle.close() # 关闭句柄(释放资源)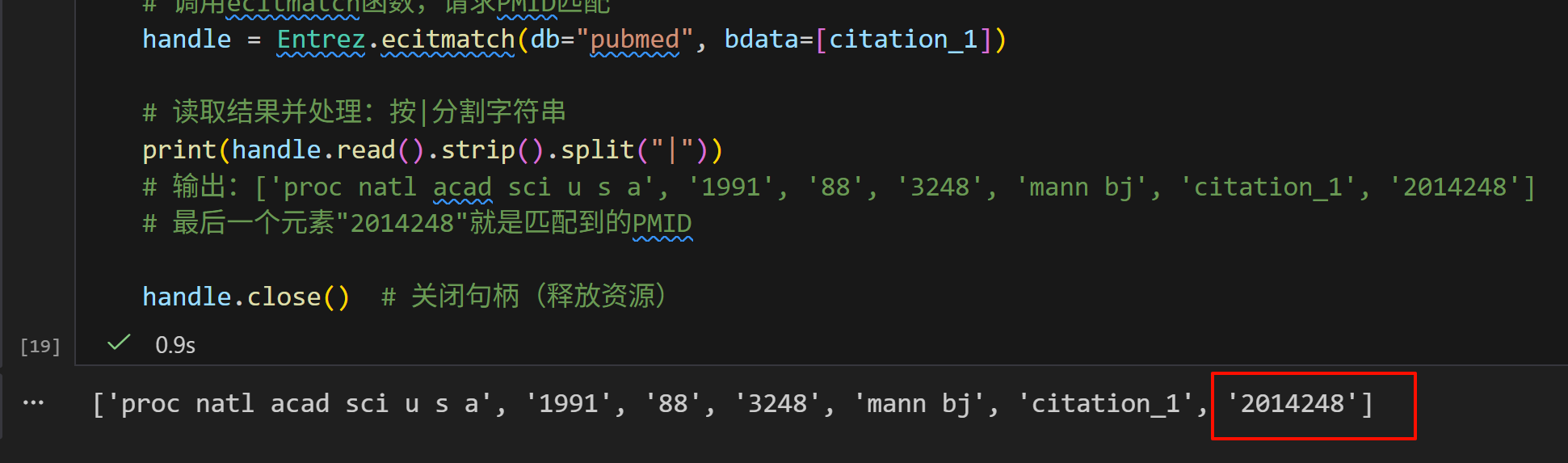
ECitMatch是 NCBI 提供的工具,专门用于 "引用信息 → PMID" 的匹配;- 适用场景:你有文献的引用要素(但不知道 PMID),需要快速定位 PubMed 中的对应文献。
- 比如说上面这个结果返回的是2014248
我们现在去验证一下,用链接:
bash
https://pubmed.ncbi.nlm.nih.gov/2014248/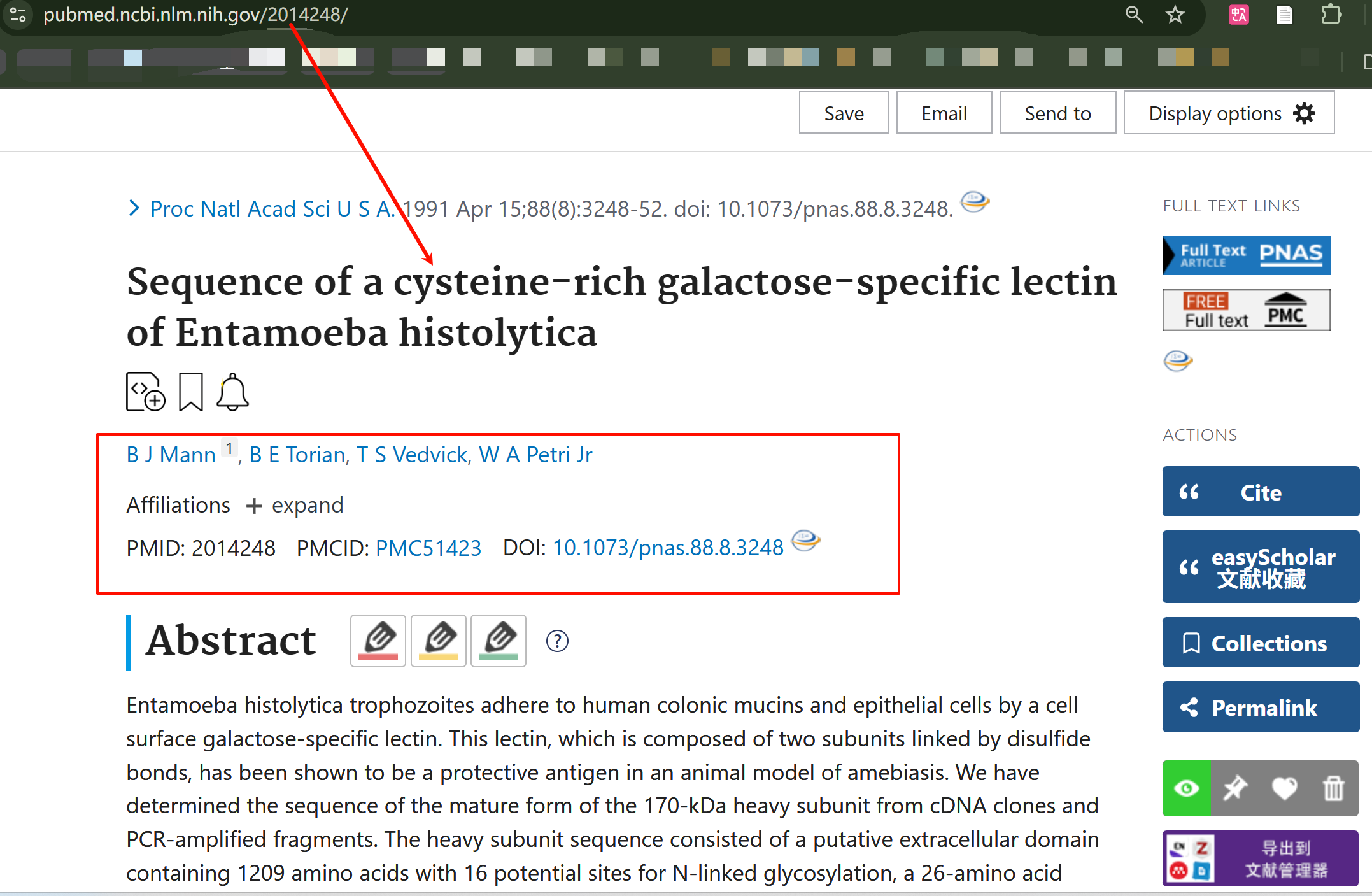
efetch
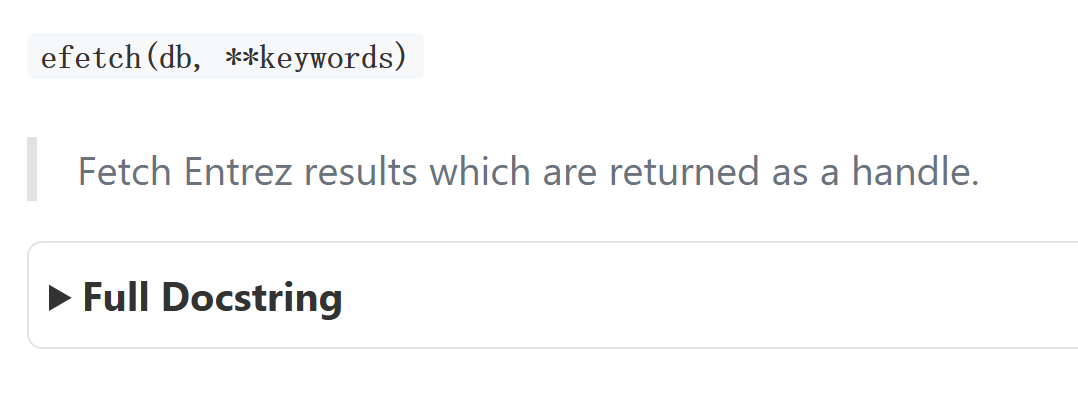

上面这段内容是对 Biopython 库中 Entrez 模块的 efetch 函数 的说明,
核心作用是:从 NCBI(美国国家生物技术信息中心)的 Entrez 数据库(如核酸、蛋白、基因库等)中,根据指定的 ID / 条件获取具体的生物信息记录,并以 "句柄(handle)" 形式返回结果,方便后续读取和处理。
| 术语 / 短语 | 概要解释 |
|---|---|
| Entrez | NCBI 提供的生物信息数据库检索系统(涵盖核酸、蛋白、文献等多类数据) |
EFetch/Entrez.efetch |
Entrez 系统的 "数据获取" 接口(对应函数),用于从 NCBI 拉取具体记录 |
| handle(句柄) | 可理解为 "数据流的操作接口",类似文件句柄,通过 .read() /.readline() 等方法读取返回的文本 / XML 数据 |
| UIs | Unique Identifiers(唯一标识符),即 NCBI 数据库中每条记录的专属 ID(如示例中的 AY851612) |
| HTTP POST/GET | 两种网络请求方式:当请求的 ID 数量超过 200 个时,函数会自动用 POST(更适合大量数据),否则用 GET |
| rettype/retmode | efetch 的核心参数:- rettype :返回数据的格式类型(如示例中 gb 代表 GenBank 格式);- retmode :返回数据的传输模式(text 文本 /xml 可扩展标记语言) |
bash
from Bio import Entrez # 导入Biopython的Entrez模块(需先安装:pip install biopython)
Entrez.email = "Your.Name.Here@example.org" # 必须填写邮箱(NCBI要求,用于追踪异常请求)
# 调用efetch获取数据:
# db="nucleotide":指定数据库为"核酸数据库"
# id="AY851612":要获取的记录ID(可传多个,如 id="AY851612,AY851613")
# rettype="gb":返回GenBank格式
# retmode="text":以纯文本形式返回
handle = Entrez.efetch(db="nucleotide", id="AY851612", rettype="gb", retmode="text")
print(handle.readline().strip()) # 读取第一行数据并去除首尾空白,输出:LOCUS AY851612 892 bp DNA linear PLN 10-APR-2007
handle.close() # 关闭句柄(释放资源)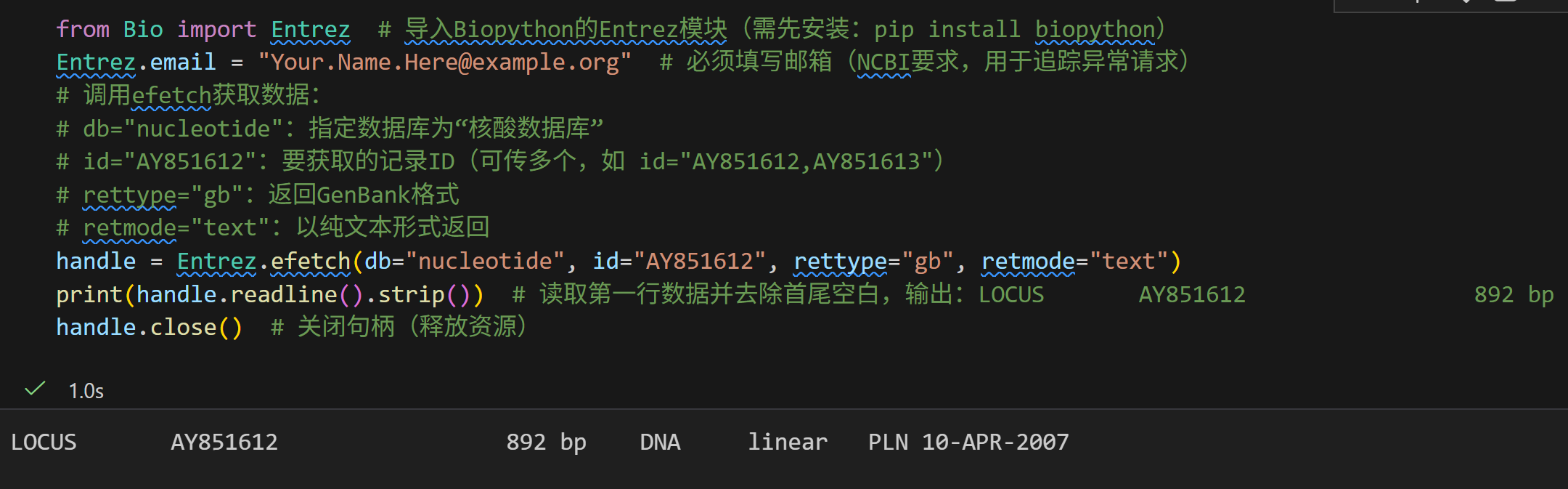
egquery(已弃用)
官方建议使用esearch
einfo
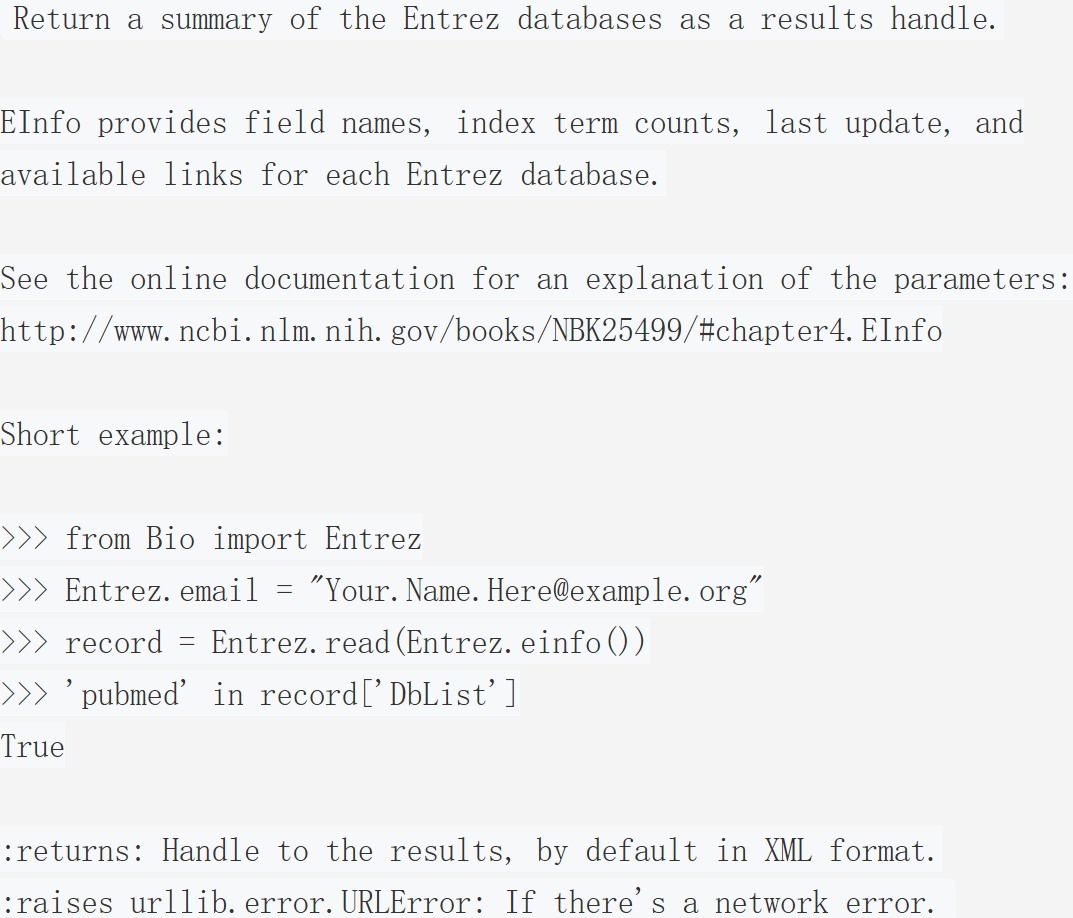
此操作核心是通过EInfo 工具获取 Entrez 数据库的摘要信息,返回的结果句柄(results handle)默认以 XML 格式呈现,包含两类关键信息:
数据库基础信息:每个 Entrez 数据库的字段名(field names)、索引术语计数(index term counts)、最后更新时间(last update)。
关联能力信息:各数据库支持的可用链接(available links),即该数据库可与其他 Entrez 数据库建立的关联关系。
elink
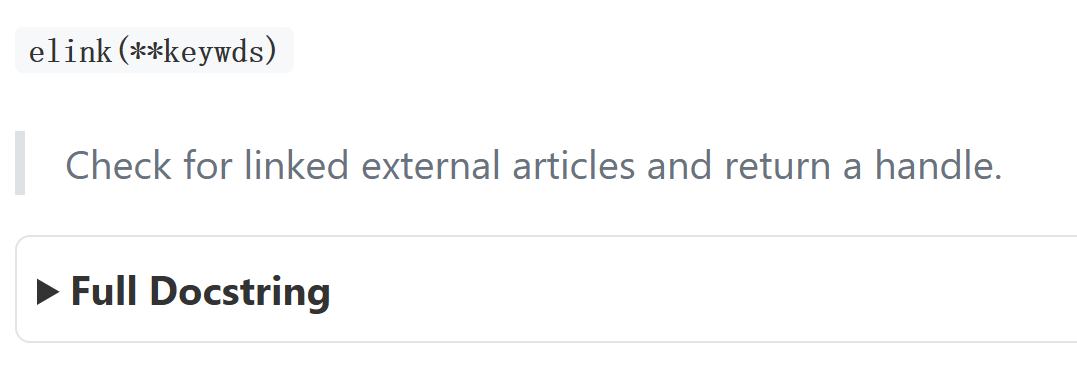


ELink 是 NCBI E-utilities 工具集中用于 "跨数据库 / 内部关联 ID 检索" 的工具,在 "外部文章 / 关联文章" 场景下,核心能力(聚焦 "外部 / 关联文章检查")包括:
- 检查链接存在性:验证一个或多个源数据库 ID(如 PubMed 的 PMID)是否存在 "外部链接(LinkOut,如期刊全文平台链接)" 或 "关联文章链接(如 PubMed 内部相关文献)"。
- 获取关联数据 :若存在链接,可检索关联的目标数据库 ID(如相关文章的 PMID)及相关性评分(仅部分模式下,如
cmd=neighbor_score)。 - 生成外部链接:为特定 ID 和数据库创建 "主要 LinkOut 提供者的直接超链接"(如某篇 PubMed 文献的期刊全文链接),或列出多个 ID 的所有 LinkOut URL 及属性(如提供者名称、链接类型)。
epost
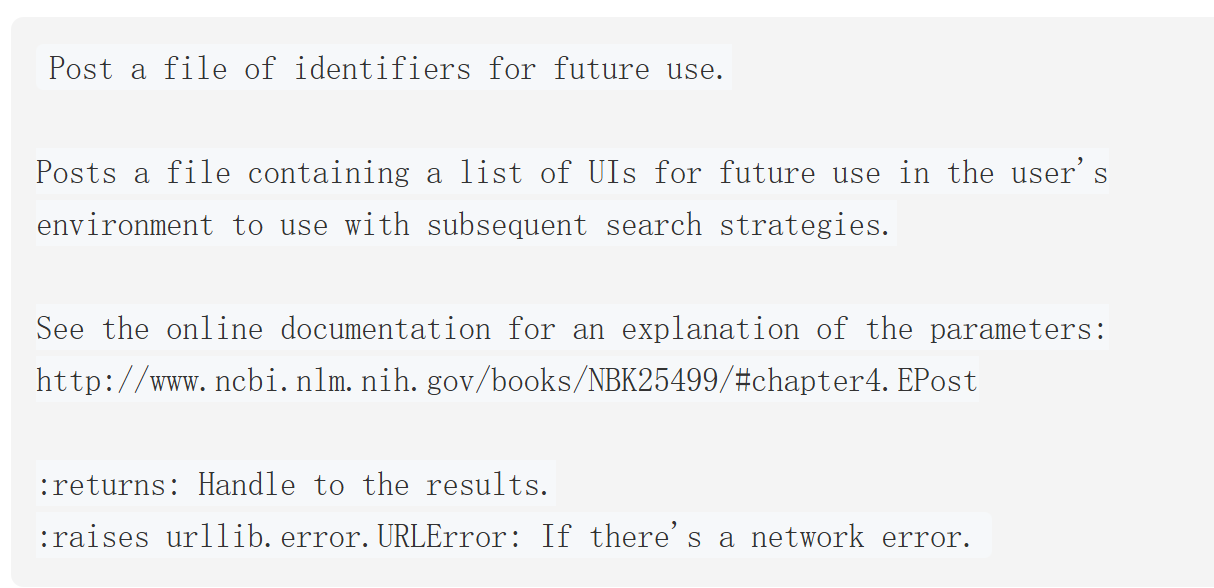
该操作核心是通过 NCBI 的EPost 工具,将包含一组标识符(UIDs,如 PubMed 的 PMID、Protein 的 GI 号或 accession.version 等)的文件上传至 Entrez History 服务器,以便后续搜索策略(如 ESearch、EFetch、ELink 等操作)直接调用这组标识符,无需重复输入,提升效率
esearch
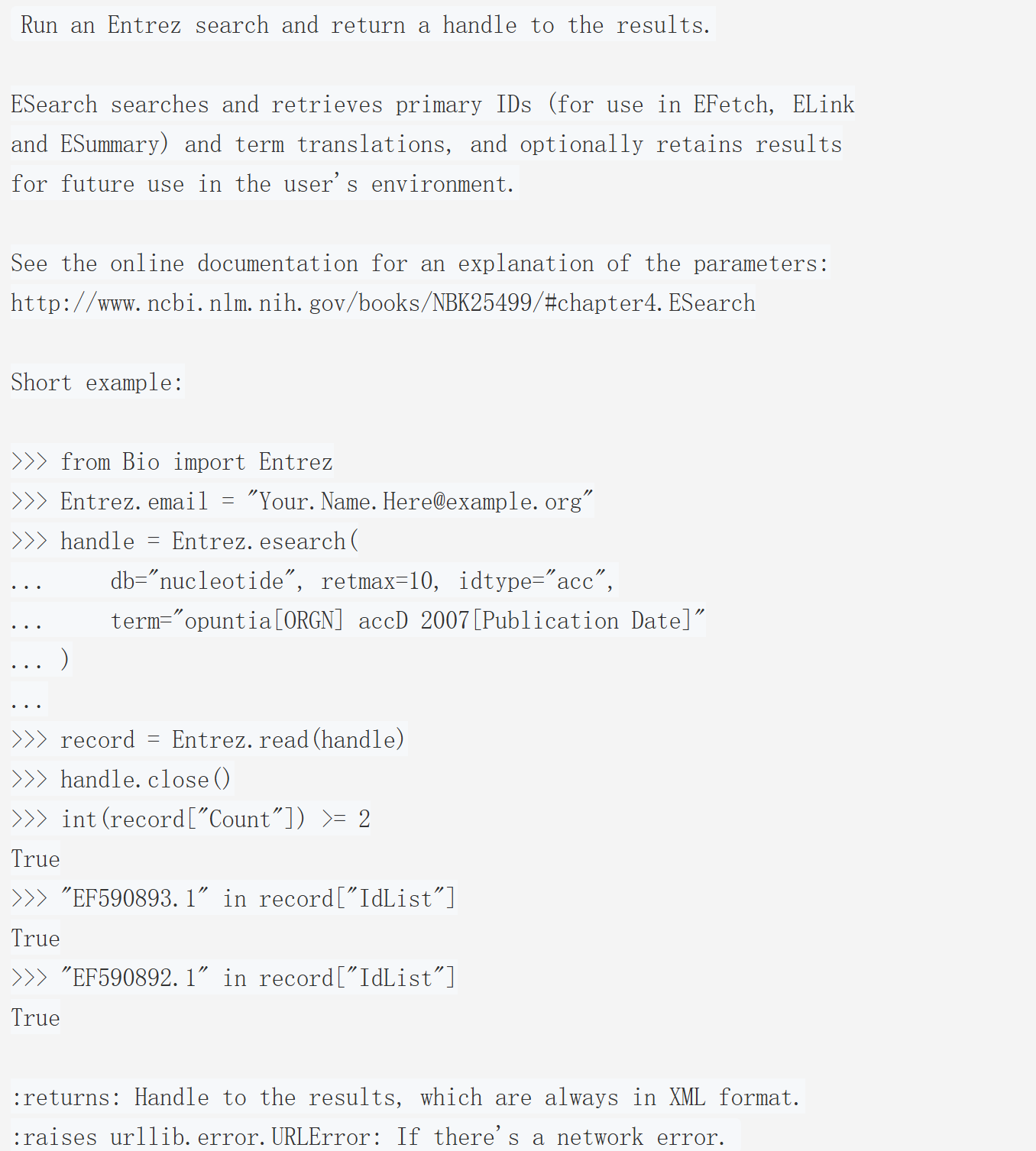
Entrez.esearch 是 Biopython 中对接 NCBI E-utilities 中 ESearch 工具的接口,核心作用是在指定的 Entrez 数据库中执行搜索,并返回可用于后续操作的结果句柄(handle)。具体能力包括:
- 检索符合查询条件的核心标识符(UID),这些 UID 可直接用于 EFetch(获取完整记录)、ELink(关联其他数据库记录)、ESummary(获取记录摘要)等后续操作;
- 返回查询词的翻译结果(如将用户输入的 "opuntia [ORGN]" 映射为数据库可识别的检索规则);
- 可选将搜索结果暂存于用户的 Entrez 环境中(通过参数控制),方便后续复用。
bash
# 1. 导入 Biopython 中操作 Entrez 的模块(没有这行,后续代码都用不了)
from Bio import Entrez
# 2. 设置你的邮箱(NCBI 强制要求!用于追踪请求来源,防止恶意访问)
# 注意:实际使用时要换成你自己的邮箱(比如 "your.email@xxx.com")
Entrez.email = "Your.Name.Here@example.org"
# 3. 调用 Entrez.esearch 执行搜索,把返回的"句柄"存在 handle 变量里
# 括号里是关键参数,每一个都对应搜索规则:
handle = Entrez.esearch(
db="nucleotide", # 指定搜索的数据库:nucleotide = 核苷酸数据库(存DNA/RNA序列)
retmax=10, # 限制返回的结果数量:最多返回10条记录(默认20条,最多1万条)
idtype="acc", # 指定返回的ID类型:acc = accession.version 格式(如 EF590893.1)
term="opuntia[ORGN] accD 2007[Publication Date]" # 搜索关键词(核心!)
)
# 这里解释下 term 关键词的逻辑(NCBI 检索格式):
# - opuntia[ORGN]:ORGN = 物种字段,意思是"只找物种为 opuntia(仙人掌属)的记录"
# - accD:直接匹配"accD 基因"(accD 是植物叶绿体中的一个功能基因)
# - 2007[Publication Date]:Publication Date = 发表日期,意思是"只找2007年发表的记录"
# 三者用空格连接,相当于"同时满足这三个条件"的记录
# 4. 用 Entrez.read() 读取"句柄"里的 XML 结果,解析成 Python 字典(record)
# 字典格式方便后续提取数据(比如总记录数、具体ID)
record = Entrez.read(handle)
# 5. 关闭"句柄"(用完要关,释放网络连接资源,避免占用)
handle.close()
# 6. 验证结果:检查符合条件的总记录数是否≥2(确保有足够结果)
# record["Count"] 是总记录数(字符串格式,所以用 int() 转成数字)
int(record["Count"]) >= 2
# 运行后返回 True,说明总记录数至少有2条
# 7. 验证结果:检查特定ID(EF590893.1、EF590892.1)是否在搜索结果里
# record["IdList"] 是本次返回的所有ID列表(比如示例中是10条里的前几条)
"EF590893.1" in record["IdList"] # 返回 True,说明这个ID在结果里
"EF590892.1" in record["IdList"] # 返回 True,说明这个ID也在结果里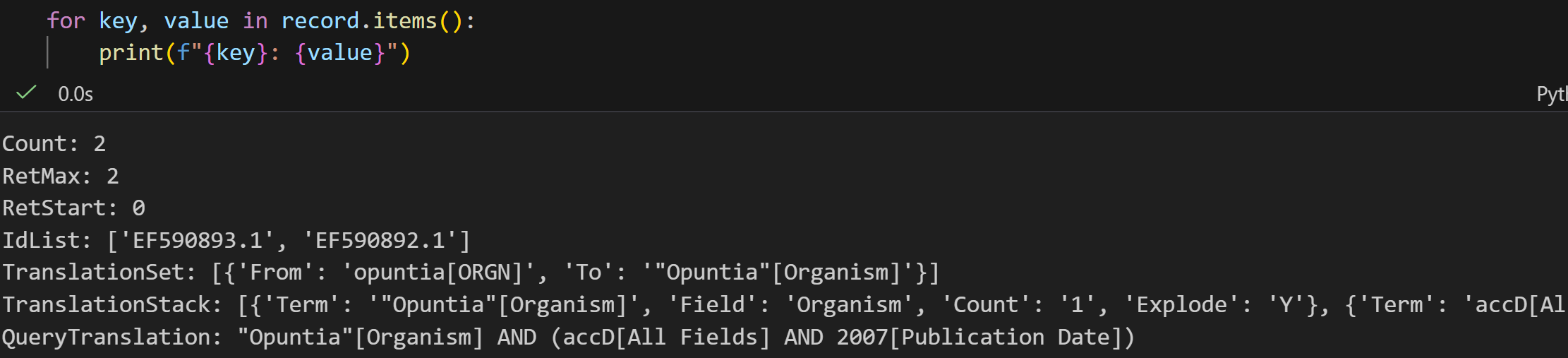
espell

ESpell 是 NCBI E-utilities 中的一个工具,核心作用是为指定数据库中的文本查询词提供拼写建议。若查询词存在拼写错误(如示例中的 "biopythooon"),它能返回纠正后的正确拼写(如 "biopython"),帮助用户优化查询以获取更准确的搜索结果。
根据链接文档,ESpell 需包含以下必填参数,无额外可选参数:
- db:目标数据库,默认值为 "pubmed"(PubMed 数据库),需传入有效的 Entrez 数据库名称。
- term:待检查拼写的 Entrez 文本查询词,特殊字符需 URL 编码,空格可替换为 "+";若查询词过长(数百字符以上),建议使用 HTTP POST 方式提交。
esummary

ESummary 是 NCBI E-utilities 中的核心工具之一,主要作用是获取文档摘要(DocSums),支持两种输入来源:
- 直接提供的一组数据库主键(UIDs,如示例中的结构数据库 ID "19923");
- 存储在 Entrez History 服务器中的 UID 集合(需配合
WebEnv和query_key参数调用)。
根据 NCBI 官方文档,ESummary 关键参数如下:
| 参数类型 | 参数名 | 说明 |
|---|---|---|
| 必选参数 | db |
目标数据库(如示例中的 "structure",即结构数据库;默认值为 "pubmed") |
| 输入来源参数 | id |
直接输入的 UID 列表(单个 ID 或逗号分隔的多个 ID,如示例中的 "19923") |
WebEnv +query_key |
从 History 服务器获取 UID 时使用,需同时提供(来自前序 ESearch/EPost 调用结果) | |
| 可选参数 | retmode |
输出格式(默认 "xml",也支持 "json") |
retmax |
单次获取的最大 DocSum 数量(上限 10000,超量需分批调用) | |
version |
若设为 "2.0",返回各数据库专属的扩展版 XML 摘要(包含更多字段) |
python
from Bio import Entrez # 导入 Biopython 的 Entrez 模块(封装 E-utilities 调用)
Entrez.email = "Your.Name.Here@example.org" # 必选:设置用户邮箱(NCBI 要求,用于合规追踪)
handle = Entrez.esummary(db="structure", id="19923") # 调用 ESummary:指定数据库为"structure",ID 为"19923"
record = Entrez.read(handle) # 解析返回的 XML 结果,转为可操作的 Python 对象
handle.close() # 关闭网络连接(避免资源泄漏)
# 提取并打印关键信息
print(record[0]["Id"]) # 输出第一个摘要的 ID(对应输入的"19923")
print(record[0]["PdbDescr"]) # 输出该结构的描述信息(大肠杆菌顺乌头酸酶 B 的晶体结构)parse
bash
Parse an XML file from the NCBI Entrez Utilities into python objects.
This function parses an XML file created by NCBI's Entrez Utilities,
returning a multilevel data structure of Python lists and dictionaries.
This function is suitable for XML files that (in Python) can be represented
as a list of individual records. Whereas 'read' reads the complete file
and returns a single Python list, 'parse' is a generator function that
returns the records one by one. This function is therefore particularly
useful for parsing large files.
Most XML files returned by NCBI's Entrez Utilities can be parsed by
this function, provided its DTD is available. Biopython includes the
DTDs for most commonly used Entrez Utilities.
The argument ``source`` must be a file or file-like object opened in binary
mode, or a filename. The parser detects the encoding from the XML file, and
uses it to convert all text in the XML to the correct Unicode string. The
functions in Bio.Entrez to access NCBI Entrez will automatically return XML
data in binary mode. For files, use mode "rb" when opening the file, as in
>>> from Bio import Entrez
>>> path = "Entrez/pubmed1.xml"
>>> stream = open(path, "rb") # opened in binary mode
>>> records = Entrez.parse(stream)
>>> for record in records:
... print(record['MedlineCitation']['Article']['Journal']['Title'])
...
Social justice (San Francisco, Calif.)
Biochimica et biophysica acta
>>> stream.close()
Alternatively, you can use the filename directly, as in
>>> records = Entrez.parse(path)
>>> for record in records:
... print(record['MedlineCitation']['Article']['Journal']['Title'])
...
Social justice (San Francisco, Calif.)
Biochimica et biophysica acta
which is safer, as the file stream will automatically be closed after all
the records have been read, or if an error occurs.
If validate is True (default), the parser will validate the XML file
against the DTD, and raise an error if the XML file contains tags that
are not represented in the DTD. If validate is False, the parser will
simply skip such tags.
If escape is True, all characters that are not valid HTML are replaced
by HTML escape characters to guarantee that the returned strings are
valid HTML fragments. For example, a less-than sign (<) is replaced by
<. If escape is False (default), the string is returned as is.
If ignore_errors is False (default), any error messages in the XML file
will raise a RuntimeError. If ignore_errors is True, error messages will
be stored as ErrorElement items, without raising an exception.
Whereas the data structure seems to consist of generic Python lists,
dictionaries, strings, and so on, each of these is actually a class
derived from the base type. This allows us to store the attributes
(if any) of each element in a dictionary my_element.attributes, and
the tag name in my_element.tag.read

bash
Parse an XML file from the NCBI Entrez Utilities into python objects.
This function parses an XML file created by NCBI's Entrez Utilities,
returning a multilevel data structure of Python lists and dictionaries.
Most XML files returned by NCBI's Entrez Utilities can be parsed by
this function, provided its DTD is available. Biopython includes the
DTDs for most commonly used Entrez Utilities.
The argument ``source`` must be a file or file-like object opened in binary
mode, or a filename. The parser detects the encoding from the XML file, and
uses it to convert all text in the XML to the correct Unicode string. The
functions in Bio.Entrez to access NCBI Entrez will automatically return XML
data in binary mode. For files, use mode "rb" when opening the file, as in
>>> from Bio import Entrez
>>> path = "Entrez/esearch1.xml"
>>> stream = open(path, "rb") # opened in binary mode
>>> record = Entrez.read(stream)
>>> print(record['QueryTranslation'])
biopython[All Fields]
>>> stream.close()
Alternatively, you can use the filename directly, as in
>>> record = Entrez.read(path)
>>> print(record['QueryTranslation'])
biopython[All Fields]
which is safer, as the file stream will automatically be closed after the
record has been read, or if an error occurs.
If validate is True (default), the parser will validate the XML file
against the DTD, and raise an error if the XML file contains tags that
are not represented in the DTD. If validate is False, the parser will
simply skip such tags.
If escape is True, all characters that are not valid HTML are replaced
by HTML escape characters to guarantee that the returned strings are
valid HTML fragments. For example, a less-than sign (<) is replaced by
<. If escape is False (default), the string is returned as is.
If ignore_errors is False (default), any error messages in the XML file
will raise a RuntimeError. If ignore_errors is True, error messages will
be stored as ErrorElement items, without raising an exception.
Whereas the data structure seems to consist of generic Python lists,
dictionaries, strings, and so on, each of these is actually a class
derived from the base type. This allows us to store the attributes
(if any) of each element in a dictionary my_element.attributes, and
the tag name in my_element.tag.parse和read的比较
基于 Biopython 中 Bio.Entrez 模块对 NCBI Entrez Utilities XML 文件的解析功能,二者的核心区别及共性如下表所示:
| 对比维度 | parse(source, validate=True, escape=False, ignore_errors=False) |
read(source, validate=True, escape=False, ignore_errors=False) |
|---|---|---|
| 返回类型与结构 | 生成器(generator),逐次返回单个记录 (如 PubMed 中的每篇文献记录),需通过循环(如 for record in records)迭代获取。 |
直接返回单个 Python 数据结构(包含整个 XML 文件的完整数据,如 eSearch 结果的全局信息),无需迭代,可直接通过键值访问。 |
| 内存占用 | 低。因生成器仅在迭代时加载当前记录,不一次性加载全部数据,适合大型 XML 文件(如包含数千条记录的 PubMed 导出文件)。 | 高。需一次性将整个 XML 文件的所有数据加载到内存,仅适合小型/单记录 XML 文件(如 eSearch、eInfo 的查询结果文件)。 |
| 适用场景 | 解析包含多个独立记录 的 XML 文件,例如: - PubMed 文献列表(pubmed.xml) - GenBank 序列批量导出文件 |
解析仅含单个全局记录的 XML 文件,例如: - eSearch 检索结果(含查询翻译、总记录数等) - eInfo 数据库信息查询结果 |
| 调用方式示例 | 生成器from Bio import Entrez; records = Entrez.parse("pubmed1.xml"); for record in records: print(record['MedlineCitation']['Article']['Title']) |
直接获取整体数据from Bio import Entrez; record = Entrez.read("esearch1.xml"); print(record['QueryTranslation']) |
| 核心功能逻辑 | 按"记录"为单位拆分 XML 数据,逐个输出,避免内存溢出。 | 将整个 XML 文件视为一个完整记录,一次性解析并返回所有层级数据(列表+字典嵌套结构)。 |
| 参数共性 | 1. source:均支持二进制模式打开的文件对象或文件名(推荐直接传文件名,自动关闭流) 2. validate:均默认验证 XML 与 DTD 一致性 3. escape:均默认不替换 HTML 特殊字符 4. ignore_errors:均默认遇到 XML 错误时抛出 RuntimeError |
同左(参数定义、默认值、功能逻辑完全一致) |
| 返回数据类型细节 | 迭代返回的每个"记录"均为 Biopython 自定义类实例(继承自列表/字典/字符串),可通过 .attributes 访问元素属性、.tag 访问标签名。 |
返回的完整数据结构同样为 Biopython 自定义类实例(继承自列表/字典),支持 .attributes 和 .tag 访问元数据。 |
| 错误处理差异 | 若 XML 中某条记录解析错误(且 ignore_errors=False),迭代到该记录时抛出错误,中断后续迭代。 |
若整个 XML 文件解析错误(且 ignore_errors=False),调用时直接抛出错误,无数据返回。 |
2,回到BioPython文档
在借助libinspector简单查看了一下文档,以及主要API之后,我们回到BioPython的Entrez这个模块。

1,EUtils XML结果的4种解析方法
EUtils(NCBI提供的编程工具集)返回的数据默认多为XML格式,需通过以下方式解析,各有适用场景:
- Bio.Entrez解析器(推荐)
Biopython专为NCBI数据设计的工具,可直接将XML结果转换为Python对象(如字典、列表),无需手动处理XML标签,操作简洁且适配NCBI数据结构,是生物信息分析中最常用的方式。 - Python标准库DOM解析器
基于"文档对象模型",会先将整个XML文件加载到内存中生成树形结构,再通过遍历树节点获取数据。适合XML文件较小、需频繁随机访问节点的场景,但内存占用较高,不适合超大文件。 - Python标准库SAX解析器
基于"事件驱动",逐行读取XML文件,遇到标签(如<DbList>)时触发预设事件(如"开始标签""结束标签")并处理数据。无需加载整个文件到内存,适合解析超大XML文件,但逻辑较复杂,需自定义事件处理函数。 - 纯文本字符串处理(不推荐)
直接将XML视为普通文本,通过字符串查找(如find()、正则表达式)提取信息。优点是无需依赖解析库,但极易受XML格式变化(如标签缩进、属性顺序)影响,稳定性差,仅适用于临时简单场景。
2,NCBI DTD文件的作用与缺失/过时处理
1. DTD文件的核心作用
DTD(文档类型定义)是NCBI用于 规范XML文件结构 的文件,定义了XML中允许的标签(如<Count>``<IdList>)、标签层级关系及数据类型。Biopython读取XML时,会通过DTD验证XML格式是否合法,确保解析结果准确。
2. DTD文件的常见问题与解决方案
Biopython默认包含多数NCBI常用DTD,但可能因NCBI升级DTD或特殊XML类型出现"缺失/过时"问题,处理方式如下:
- 自动补救 :若DTD缺失,
Entrez.read()会弹出警示(含缺失DTD的名称和下载URL),并自动从互联网下载DTD,保证解析继续进行(但会增加网络耗时)。 - 手动优化(提升速度) :为避免重复联网下载,可手动将DTD文件放入指定目录,解析器会优先读取本地文件:
- 优先目录:用户主目录下的
~/.biopython/Bio/Entrez/DTDs(无权限操作系统目录时首选); - 系统目录:
.../site-packages/Bio/Entrez/DTDs(Biopython安装目录,需管理员权限); - 源码安装场景:将DTD放入Biopython源码的
Bio/Entrez/DTDs目录,重新安装即可覆盖旧文件。
- 优先目录:用户主目录下的
简单来说就是EUtils工具返回的大多是XML格式数据,DTD是NCBI用来规范、描述这个返回的XML格式的文件,而Bio.Entrez解析器能够解析这个返回的XML格式数据。(简单来说就是NCBI有一大堆数据(XML)要返回,DTD是其说明文档/指南,解析器是直接处理这些数据的)。
Entrez Programming Utilities也可以生成其他格式的输出文件,比如Fasta、序列数据库里面的GenBank文件格式 或者文献数据库里面的MedLine格式。
3,通过Biopython访问NCBI的线上资源规范(NCBI的Entrez 用户规范)
如果NCBI发现你在滥用他们的系统,他们会禁止你的访问。

最后,根据你的使用情况选择不同的策略。如果你打算下载大量的数据,最好使用其他的方法。比如,你想得到所有人的 基因的数据,那么考虑通过FTP得到每个染色体的GenBank文件,然后将这些文件导入到你自己的BioSQL数据库里面去。
4,EInfo: 获取Entrez数据库的信息
EInfo为每个NCBI的数据库提供了条目索引,最近更新的时间以及可用的链接。此外,你可以很容易的使用EInfo通过 Entrez获取所有数据库名字的列表:
python
from Bio import Entrez
Entrez.email = "A.N.Other@example.com" # Always tell NCBI who you are
handle = Entrez.einfo()
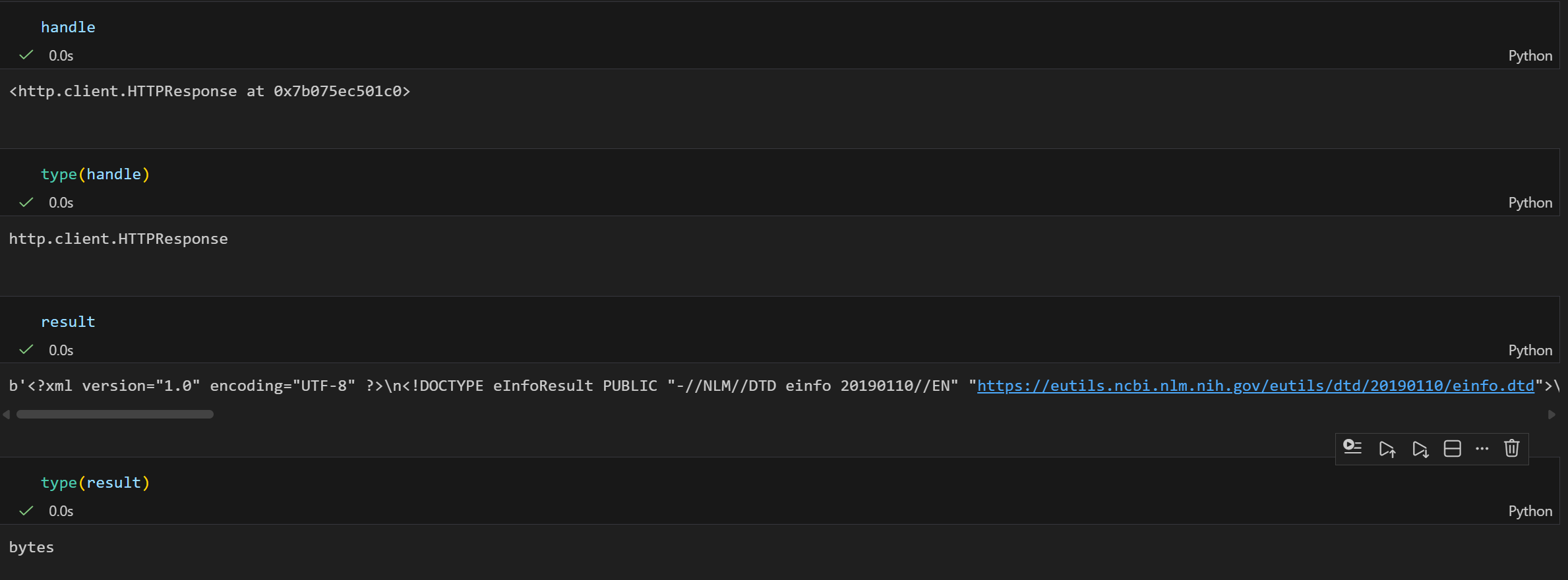
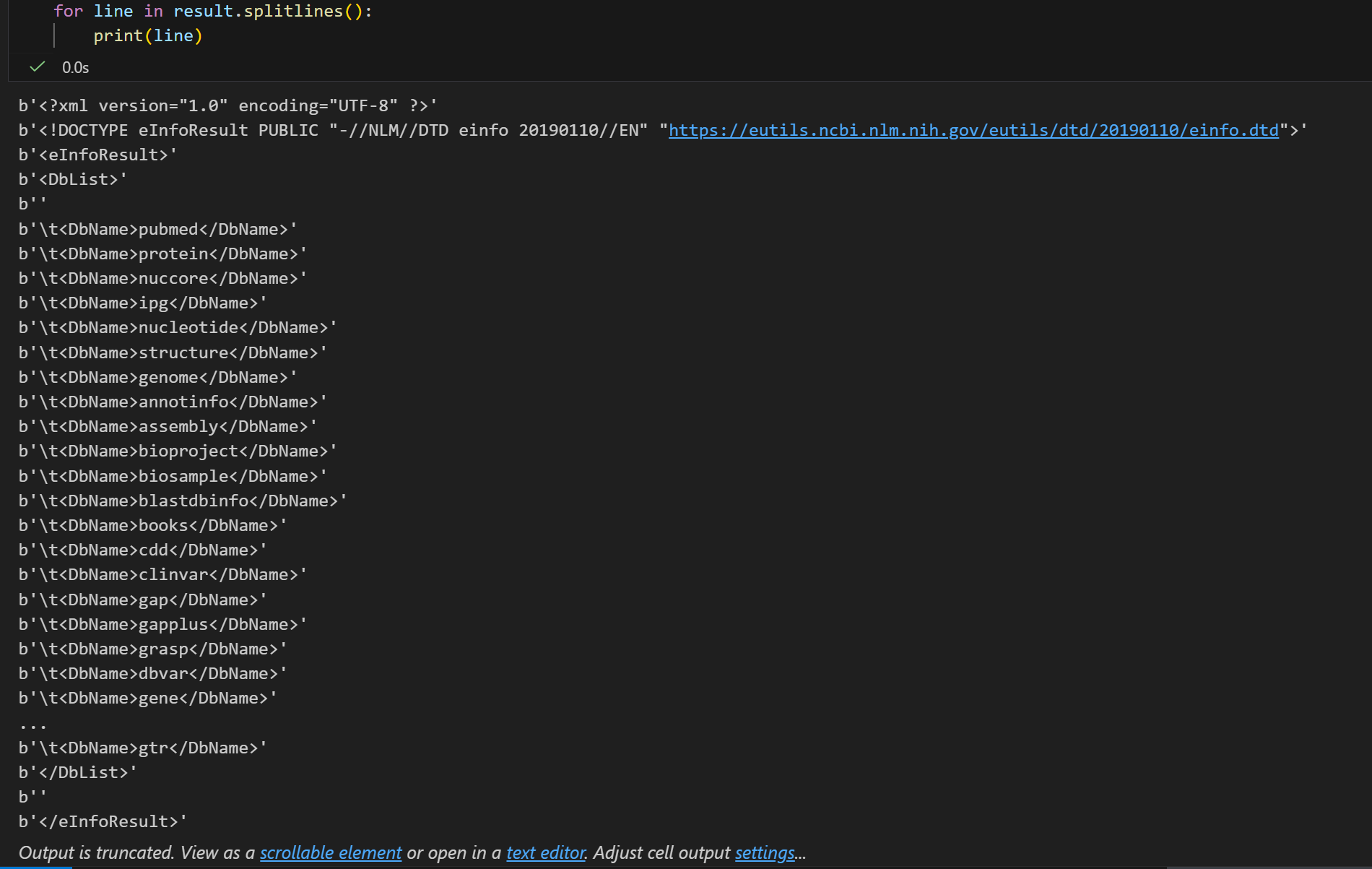
result = handle.read()依据下面的内容,我们基本上可以确定:
- type(handle) → http.client.HTTPResponse(http响应对象):Bio.Entrez 库的核心是向 NCBI 的 Entrez 接口发送 HTTP 请求,而 einfo() 方法执行后,返回的 handle 本质是 Python 标准库 http.client.HTTPResponse 类型的对象,它代表的是服务器返回的原始 HTTP 响应流,而不是直接的 XML 文本。
- type(result) → str(XML 格式的文本字符串,响应内容/XML格式):当我们调用 handle.read() 时,会读取这个 HTTP 响应流里的字节数据,并解码为字符串,这个字符串的内容就是 NCBI 返回的 XML 格式数据。
现在我们来看看这个http响应的具体内容,原始是XML格式的(html其实也是XML格式的一种特殊形式,所以熟悉Python爬虫的看这些应该都不陌生)。
变量 result 现在包含了XML格式的数据库列表:
因为有换行符,所以我们不直接用print,还是用splitlines按照换行符拆分
python
for line in result.splitlines():
print(line)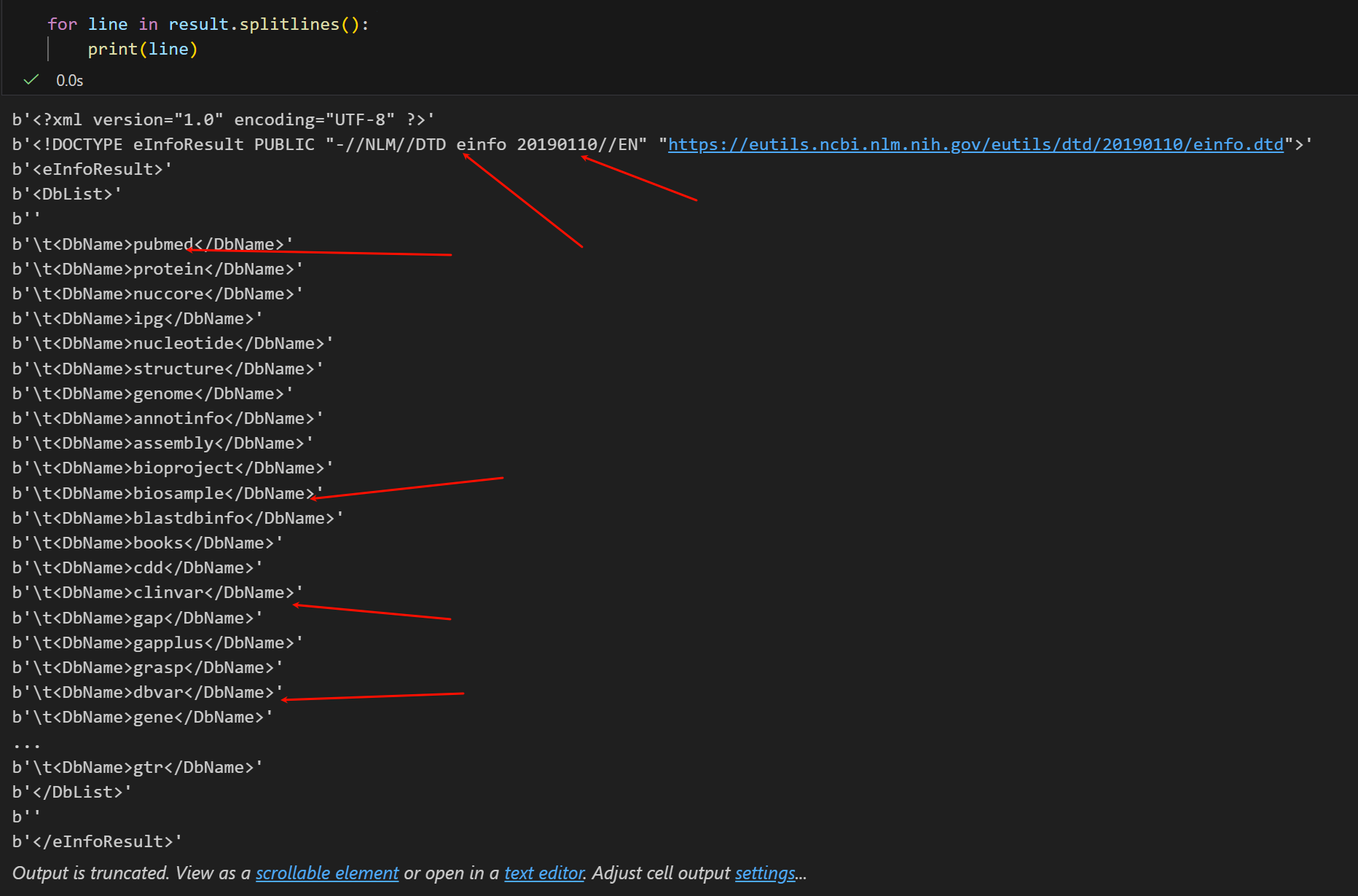
返回的XML格式中(我们可以类比特殊的html来观察分析),
像各种数据库我们就不用说了pubmed基本上生物医学领域的没有人不知道,clivar也是常用的临床数据库,等等等等。
其中DTD不用说了,我们前面提到过,相当于是使用文档/说明书,我们直接点开右边这个链接就能够拿到这个DTD文件
python
<!--
This is the Current DTD for Entrez eInfo
$Id: einfo.dtd 577924 2019-01-09 22:59:07Z fialkov $
need xml2json.xsl
-->
<!-- ================================================================= -->
<!--~~ !dtd
~~json
<json type='einfo' version='0.3'>
<config lcnames='true'/>
</json>
~~-->
<!ELEMENT DbName (#PCDATA)> <!-- \S+ -->
<!ELEMENT Name (#PCDATA)> <!-- .+ -->
<!ELEMENT FullName (#PCDATA)> <!-- .+ -->
<!ELEMENT Description (#PCDATA)> <!-- .+ -->
<!ELEMENT DbBuild (#PCDATA)> <!-- .+ -->
<!ELEMENT TermCount (#PCDATA)> <!-- \d+ -->
<!ELEMENT Menu (#PCDATA)> <!-- .+ -->
<!ELEMENT DbTo (#PCDATA)> <!-- \S+ -->
<!ELEMENT MenuName (#PCDATA)> <!-- .+ -->
<!ELEMENT Count (#PCDATA)> <!-- \d+ -->
<!ELEMENT LastUpdate (#PCDATA)> <!-- \d+ -->
<!ELEMENT ERROR (#PCDATA)> <!-- .+ -->
<!ELEMENT Warning (#PCDATA)> <!-- .+ -->
<!ELEMENT IsDate (#PCDATA)> <!-- (Y|N) -->
<!ELEMENT IsNumerical (#PCDATA)> <!-- (Y|N) -->
<!ELEMENT SingleToken (#PCDATA)> <!-- (Y|N) -->
<!ELEMENT Hierarchy (#PCDATA)> <!-- (Y|N) -->
<!ELEMENT IsHidden (#PCDATA)> <!-- (Y|N) -->
<!ELEMENT IsRangable (#PCDATA)> <!-- (Y|N) -->
<!ELEMENT IsTruncatable (#PCDATA)> <!-- (Y|N) -->
<!ELEMENT DbList (DbName+)>
<!ELEMENT Field (Name,
FullName,
Description,
TermCount,
IsDate,
IsNumerical,
SingleToken,
Hierarchy,
IsHidden,
IsRangable?,
IsTruncatable?)>
<!ELEMENT Link (Name,Menu,Description,DbTo)>
<!ELEMENT LinkList (Link*)>
<!ELEMENT FieldList (Field*)>
<!ELEMENT DbInfo (DbName,
MenuName,
Description,
DbBuild?,
Warning?,
Count?,
LastUpdate?,
FieldList?,
LinkList?)>
<!--~~ <eInfoResult>
~~ json
<object>
<member select='DbList'/>
<array key='dbinfo' select='DbInfo|ERROR'/>
</object>
~~-->
<!ELEMENT eInfoResult (DbList|(DbInfo|ERROR)+)>当然,这个数据我们也可以直接从biopython本身的package文件中查找:前面说了是在...site-packages/Bio/Entrez/DTDs中。
我们的biopython版本比较新了,是1.85,但是这个DTD还是2019年的,只能说数据库就这样了(其实没怎么大更新过,我之前其实还担心biopython项目太大,维护不及时那就很麻烦了)
可以看到,这个文件夹里面有非常多的DTD文件,说明每一个数目库返回的格式都是不一样不统一的,都需要单独额外进行规划:
另外我红圈标注出来的是文献数据库Pubmed
还有一大堆:

我这里把最后一个190101的dtd拉出来,大家可以看一下:
只能说文献数据库本身也是比较复杂的
python
<!--
2018-09-01
This DTD supports both the E-utilities and ftp service data dissemination methods.
It is based on http://dtd.nlm.nih.gov/ncbi/pubmed/out/pubmed_190101.dtd
Additions/Changes since 180601 DTD:
1. Added elements to capture reference citations:
<ReferenceList>
<Reference>
<Citation>
2. Removed <CitationString> from book records.
3. Added four values to CommentsCorrections/@RefType
CorrectedandRepublishedIn
CorrectedandRepublishedFrom
RetractedandRepublishedIn
RetractedandRepublishedFrom
4. Added "plain-language-summary" to allowed values for @Type on
<OtherAbstract>
NOTE: The use of "Medline" in a DTD or element name does not mean the record
represents a citation from a MEDLINE-selected journal. When the NLM DTDs and
XML elements were first created, MEDLINE records were the only data exported.
Now NLM exports citations other than MEDLINE records using these tools. To
minimize unnecessary disruption to users of the data and tools, NLM has
retained the original DTD and element names (e.g., MedlineTA, MedlineJournalInfo).
NOTE: StartPage and EndPage in Pagination element are not currently used; are
reserved for future use.
* = 0 or more occurrences (optional element, repeatable)
? = 0 or 1 occurrences (optional element, at most 1)
+ = 1 or more occurrences (required element, repeatable)
| = choice, one or the other but not both
no symbol = required element
-->
<!-- ============================================================= -->
<!-- MATHML 3.0 SETUP -->
<!-- ============================================================= -->
<!-- MATHML SETUP FILE -->
<!ENTITY % mathml-in-pubmed SYSTEM "mathml-in-pubmed.mod" >
%mathml-in-pubmed;
<!-- ================================================================= -->
<!-- ================================================================= -->
<!ENTITY % text "#PCDATA | b | i | sup | sub | u" >
<!ENTITY % booklinkatts
"book CDATA #IMPLIED
part CDATA #IMPLIED
sec CDATA #IMPLIED" >
<!-- ================================================================= -->
<!-- ================================================================= -->
<!-- ================= Set-level elements ============================-->
<!ELEMENT PubmedArticleSet ((PubmedArticle | PubmedBookArticle)+, DeleteCitation?) >
<!ATTLIST PubmedArticleSet
>
<!ELEMENT BookDocumentSet (BookDocument*, DeleteDocument?) >
<!ATTLIST BookDocumentSet
>
<!ELEMENT PubmedBookArticleSet (PubmedBookArticle*)>
<!ATTLIST PubmedBookArticleSet
>
<!-- ============= Document-level elements ============================-->
<!ELEMENT PubmedArticle (MedlineCitation, PubmedData?)>
<!ATTLIST PubmedArticle
>
<!ELEMENT PubmedBookArticle (BookDocument, PubmedBookData?)>
<!ATTLIST PubmedBookArticle
>
<!ELEMENT BookDocument ( PMID, ArticleIdList, Book, LocationLabel*, ArticleTitle?, VernacularTitle?,
Pagination?, Language*, AuthorList*, InvestigatorList?, PublicationType*, Abstract?, Sections?, KeywordList*,
ContributionDate?, DateRevised?, GrantList?, ItemList*, ReferenceList*) >
<!ELEMENT DeleteCitation (PMID+) >
<!ELEMENT DeleteDocument (PMID*) >
<!-- =============== Sub-Document wrapper elements =====================-->
<!ELEMENT MedlineCitation (PMID, DateCompleted?, DateRevised?, Article,
MedlineJournalInfo, ChemicalList?, SupplMeshList?,CitationSubset*,
CommentsCorrectionsList?, GeneSymbolList?, MeshHeadingList?,
NumberOfReferences?, PersonalNameSubjectList?, OtherID*, OtherAbstract*,
KeywordList*, CoiStatement?, SpaceFlightMission*, InvestigatorList?, GeneralNote*)>
<!ATTLIST MedlineCitation
Owner (NLM | NASA | PIP | KIE | HSR | HMD | NOTNLM) "NLM"
Status (Completed | In-Process | PubMed-not-MEDLINE | In-Data-Review | Publisher |
MEDLINE | OLDMEDLINE) #REQUIRED
VersionID CDATA #IMPLIED
VersionDate CDATA #IMPLIED
IndexingMethod CDATA #IMPLIED >
<!ELEMENT PubmedData (History?, PublicationStatus, ArticleIdList, ObjectList?, ReferenceList*) >
<!ELEMENT PubmedBookData (History?, PublicationStatus, ArticleIdList, ObjectList?)>
<!ELEMENT Article (Journal,ArticleTitle,((Pagination, ELocationID*) | ELocationID+),
Abstract?,AuthorList?, Language+, DataBankList?, GrantList?,
PublicationTypeList, VernacularTitle?, ArticleDate*) >
<!ATTLIST Article
PubModel (Print | Print-Electronic | Electronic | Electronic-Print | Electronic-eCollection) #REQUIRED >
<!-- ================================================================= -->
<!-- Everything else in alphabetical order -->
<!-- ================================================================= -->
<!ELEMENT Abstract (AbstractText+, CopyrightInformation?)>
<!ELEMENT AbstractText (%text; | mml:math | DispFormula)* >
<!ATTLIST AbstractText
Label CDATA #IMPLIED
NlmCategory (BACKGROUND | OBJECTIVE | METHODS | RESULTS | CONCLUSIONS | UNASSIGNED) #IMPLIED >
<!ELEMENT AccessionNumber (#PCDATA) >
<!ELEMENT AccessionNumberList (AccessionNumber+) >
<!ELEMENT Acronym (#PCDATA) >
<!ELEMENT Affiliation (%text;)*>
<!ELEMENT AffiliationInfo (Affiliation, Identifier*)>
<!ELEMENT Agency (#PCDATA) >
<!ELEMENT ArticleDate (Year, Month, Day) >
<!ATTLIST ArticleDate
DateType CDATA #FIXED "Electronic" >
<!ELEMENT ArticleId (#PCDATA) >
<!ATTLIST ArticleId
IdType (doi | pii | pmcpid | pmpid | pmc | mid |
sici | pubmed | medline | pmcid | pmcbook | bookaccession) "pubmed" >
<!ELEMENT ArticleIdList (ArticleId+)>
<!ELEMENT ArticleTitle (%text; | mml:math)*>
<!ATTLIST ArticleTitle %booklinkatts; >
<!ELEMENT Author (((LastName, ForeName?, Initials?, Suffix?) | CollectiveName), Identifier*, AffiliationInfo*) >
<!ATTLIST Author
ValidYN (Y | N) "Y"
EqualContrib (Y | N) #IMPLIED >
<!ELEMENT AuthorList (Author+) >
<!ATTLIST AuthorList
CompleteYN (Y | N) "Y"
Type ( authors | editors ) #IMPLIED >
<!ELEMENT b (%text;)*> <!-- bold -->
<!ELEMENT BeginningDate ( Year, ((Month, Day?) | Season)? ) >
<!ELEMENT Book ( Publisher, BookTitle, PubDate, BeginningDate?, EndingDate?, AuthorList*, InvestigatorList?, Volume?,
VolumeTitle?, Edition?, CollectionTitle?, Isbn*, ELocationID*, Medium?, ReportNumber?) >
<!ELEMENT BookTitle (%text; | mml:math)*>
<!ATTLIST BookTitle %booklinkatts; >
<!ELEMENT Chemical (RegistryNumber, NameOfSubstance) >
<!ELEMENT ChemicalList (Chemical+) >
<!ELEMENT Citation (%text; | mml:math)*>
<!ELEMENT CitationSubset (#PCDATA) >
<!ELEMENT CoiStatement (%text;)*>
<!ELEMENT CollectionTitle (%text; | mml:math)*>
<!ATTLIST CollectionTitle %booklinkatts; >
<!ELEMENT CollectiveName (%text;)*>
<!ELEMENT CommentsCorrections (RefSource,PMID?,Note?) >
<!ATTLIST CommentsCorrections
RefType (AssociatedDataset |
AssociatedPublication |
CommentIn | CommentOn |
CorrectedandRepublishedIn | CorrectedandRepublishedFrom |
ErratumIn | ErratumFor |
ExpressionOfConcernIn | ExpressionOfConcernFor |
RepublishedIn | RepublishedFrom |
RetractedandRepublishedIn | RetractedandRepublishedFrom |
RetractionIn | RetractionOf |
UpdateIn | UpdateOf |
SummaryForPatientsIn |
OriginalReportIn |
ReprintIn | ReprintOf |
Cites) #REQUIRED >
<!ELEMENT CommentsCorrectionsList (CommentsCorrections+) >
<!ELEMENT ContractNumber (#PCDATA) >
<!ELEMENT ContributionDate ( Year, ((Month, Day?) | Season)? ) >
<!ELEMENT CopyrightInformation (#PCDATA) >
<!ELEMENT Country (#PCDATA) >
<!ELEMENT DataBank (DataBankName, AccessionNumberList?) >
<!ELEMENT DataBankList (DataBank+) >
<!ATTLIST DataBankList
CompleteYN (Y | N) "Y" >
<!ELEMENT DataBankName (#PCDATA) >
<!ELEMENT DateCompleted (Year,Month,Day) >
<!ELEMENT DateRevised (Year,Month,Day) >
<!ELEMENT Day (#PCDATA )>
<!ELEMENT DescriptorName (#PCDATA) >
<!ATTLIST DescriptorName
MajorTopicYN (Y | N) "N"
Type (Geographic) #IMPLIED
UI CDATA #REQUIRED >
<!ELEMENT DispFormula (mml:math) >
<!ELEMENT Edition (#PCDATA) >
<!ELEMENT ELocationID (#PCDATA) >
<!ATTLIST ELocationID
EIdType (doi | pii) #REQUIRED
ValidYN (Y | N) "Y">
<!ELEMENT EndingDate ( Year, ((Month, Day?) | Season)? ) >
<!ELEMENT EndPage (#PCDATA) >
<!ELEMENT ForeName (#PCDATA) >
<!ELEMENT GeneSymbol (#PCDATA) >
<!ELEMENT GeneSymbolList (GeneSymbol+)>
<!ELEMENT GeneralNote (#PCDATA) >
<!ATTLIST GeneralNote
Owner (NLM | NASA | PIP | KIE | HSR | HMD) "NLM" >
<!ELEMENT Grant (GrantID?, Acronym?, Agency, Country)>
<!ELEMENT GrantID (#PCDATA) >
<!ELEMENT GrantList (Grant+)>
<!ATTLIST GrantList
CompleteYN (Y | N) "Y">
<!ELEMENT History (PubMedPubDate+) >
<!ELEMENT Hour (#PCDATA) >
<!ELEMENT i (%text;)*> <!-- italic -->
<!ELEMENT Identifier (#PCDATA) >
<!ATTLIST Identifier
Source CDATA #REQUIRED >
<!ELEMENT Initials (#PCDATA) >
<!ELEMENT Investigator (LastName, ForeName?, Initials?, Suffix?, Identifier*, AffiliationInfo*) >
<!ATTLIST Investigator
ValidYN (Y | N) "Y" >
<!ELEMENT InvestigatorList (Investigator+) >
<!ELEMENT Isbn (#PCDATA) >
<!ELEMENT ISOAbbreviation (#PCDATA) >
<!ELEMENT ISSN (#PCDATA) >
<!ATTLIST ISSN
IssnType (Electronic | Print) #REQUIRED >
<!ELEMENT ISSNLinking (#PCDATA) >
<!ELEMENT Issue (#PCDATA) >
<!ELEMENT Item (#PCDATA)>
<!ELEMENT ItemList (Item+)>
<!ATTLIST ItemList
ListType CDATA #REQUIRED>
<!ELEMENT Journal (ISSN?, JournalIssue, Title?, ISOAbbreviation?)>
<!ELEMENT JournalIssue (Volume?, Issue?, PubDate) >
<!ATTLIST JournalIssue
CitedMedium (Internet | Print) #REQUIRED >
<!ELEMENT Keyword (%text; | mml:math)*>
<!ATTLIST Keyword
MajorTopicYN (Y | N) "N" >
<!ELEMENT KeywordList (Keyword+) >
<!ATTLIST KeywordList
Owner (NLM | NLM-AUTO | NASA | PIP | KIE | NOTNLM | HHS) "NLM" >
<!ELEMENT Language (#PCDATA) >
<!ELEMENT LastName (#PCDATA) >
<!ELEMENT LocationLabel (#PCDATA)>
<!ATTLIST LocationLabel
Type (part|chapter|section|appendix|figure|table|box) #IMPLIED >
<!ELEMENT Medium (#PCDATA) >
<!ELEMENT MedlineDate (#PCDATA) >
<!ELEMENT MedlineJournalInfo (Country?, MedlineTA, NlmUniqueID?, ISSNLinking?) >
<!ELEMENT MedlinePgn (#PCDATA) >
<!ELEMENT MedlineTA (#PCDATA) >
<!ELEMENT MeshHeading (DescriptorName, QualifierName*)>
<!ELEMENT MeshHeadingList (MeshHeading+)>
<!ELEMENT Minute (#PCDATA) >
<!ELEMENT Month (#PCDATA) >
<!ELEMENT NameOfSubstance (#PCDATA) >
<!ATTLIST NameOfSubstance
UI CDATA #REQUIRED >
<!ELEMENT NlmUniqueID (#PCDATA) >
<!ELEMENT Note (#PCDATA) >
<!ELEMENT NumberOfReferences (#PCDATA) >
<!ELEMENT Object (Param*)>
<!ATTLIST Object
Type CDATA #REQUIRED >
<!ELEMENT ObjectList (Object+) >
<!ELEMENT OtherAbstract (AbstractText+, CopyrightInformation?) >
<!ATTLIST OtherAbstract
Type (AAMC | AIDS | KIE | PIP | NASA | Publisher |
plain-language-summary) #REQUIRED
Language CDATA "eng" >
<!ELEMENT OtherID (#PCDATA) >
<!ATTLIST OtherID
Source (NASA | KIE | PIP | POP | ARPL | CPC | IND | CPFH | CLML |
NRCBL | NLM | QCIM) #REQUIRED >
<!ELEMENT PMID (#PCDATA) >
<!ATTLIST PMID
Version CDATA #REQUIRED >
<!ELEMENT Pagination ((StartPage, EndPage?, MedlinePgn?) | MedlinePgn) >
<!ELEMENT Param (%text;)*>
<!ATTLIST Param
Name CDATA #REQUIRED >
<!ELEMENT PersonalNameSubject (LastName, ForeName?, Initials?, Suffix?) >
<!ELEMENT PersonalNameSubjectList (PersonalNameSubject+) >
<!ELEMENT PubDate ((Year, ((Month, Day?) | Season)?) | MedlineDate) >
<!ELEMENT PublicationStatus (#PCDATA) >
<!ELEMENT PublicationType (#PCDATA) >
<!ATTLIST PublicationType
UI CDATA #REQUIRED >
<!ELEMENT PublicationTypeList (PublicationType+) >
<!ELEMENT PubMedPubDate (Year, Month, Day, (Hour, (Minute, Second?)?)?)>
<!ATTLIST PubMedPubDate
PubStatus (received | accepted | epublish |
ppublish | revised | aheadofprint |
retracted | ecollection | pmc | pmcr | pubmed | pubmedr |
premedline | medline | medliner | entrez | pmc-release) #REQUIRED >
<!ELEMENT Publisher (PublisherName, PublisherLocation?) >
<!ELEMENT PublisherLocation (#PCDATA) >
<!ELEMENT PublisherName (%text;)*>
<!ELEMENT QualifierName (#PCDATA) >
<!ATTLIST QualifierName
MajorTopicYN (Y | N) "N"
UI CDATA #REQUIRED >
<!ELEMENT Reference (Citation, ArticleIdList?) >
<!ELEMENT ReferenceList (Title?, Reference*, ReferenceList*) >
<!ELEMENT RefSource (#PCDATA) >
<!ELEMENT RegistryNumber (#PCDATA) >
<!ELEMENT ReportNumber (#PCDATA) >
<!ELEMENT Season (#PCDATA) >
<!ELEMENT Second (#PCDATA) >
<!ELEMENT Section (LocationLabel?, SectionTitle, Section*) >
<!ELEMENT Sections (Section+) >
<!ELEMENT SectionTitle (%text;)*>
<!ATTLIST SectionTitle %booklinkatts; >
<!ELEMENT SpaceFlightMission (#PCDATA) >
<!ELEMENT StartPage (#PCDATA) >
<!ELEMENT sub (%text;)*> <!-- subscript -->
<!ELEMENT Suffix (%text;)*>
<!ELEMENT sup (%text;)*> <!-- superscript -->
<!ELEMENT SupplMeshList (SupplMeshName+)>
<!ELEMENT SupplMeshName (#PCDATA) >
<!ATTLIST SupplMeshName
Type (Disease | Protocol | Organism) #REQUIRED
UI CDATA #REQUIRED >
<!ELEMENT Title (#PCDATA) >
<!ELEMENT u (%text;)*> <!-- underline -->
<!ELEMENT URL (#PCDATA) >
<!ATTLIST URL
lang (AF|AR|AZ|BG|CS|DA|DE|EN|EL|ES|FA|FI|FR|HE|
HU|HY|IN|IS|IT|IW|JA|KA|KO|LT|MK|ML|NL|NO|
PL|PT|PS|RO|RU|SL|SK|SQ|SR|SV|SW|TH|TR|UK|
VI|ZH) #IMPLIED
Type ( FullText | Summary | fulltext | summary) #IMPLIED >
<!ELEMENT VernacularTitle (%text; | mml:math)*>
<!ELEMENT Volume (#PCDATA) >
<!ELEMENT VolumeTitle (%text;)*>
<!ELEMENT Year (#PCDATA) >从上面内容中可以看出,其实Einfo本质上就是查询 Entrez 系统中可用数据库的 "元信息"------ 比如有哪些数据库、每个数据库的字段(Field)含义、可关联的其他数据库(Link)、数据更新时间等,相当于 Entrez 系统的 "说明书查询工具"。
然后上面其实是两部分内容, einfo.dtd(XML 语法规则,也就是我们点击下载的那个链接文件)和 eInfoResult(实际返回的 XML 数据,也就是我们将einfo的result进行print的内容),二者是 "规则 - 实例" 的关系。
因为这两个比较重要,是我们理解NCBI等数据库底层组织以及构建的重点,在我的理解中NCBI就是一个链接型(link type)数据库,我们再详细讲一下:
1. einfo.dtd:定义XML的结构规则
DTD(文档类型定义)的作用是约束XML文件的标签、层级和数据格式,确保所有eInfo返回的XML都符合统一标准。以下是关键标签的含义(按功能分类):
| 标签组 | 核心标签 | 含义与约束 |
|---|---|---|
| 基础信息标签 | DbName、Name、FullName | - DbName:数据库唯一标识(如pubmed、protein,必须是无空格字符串) - Name:字段/链接的简称 - FullName:字段/数据库的全称(如"PubMed") |
| 数据库元信息 | DbInfo、DbBuild、Count | - DbInfo:单个数据库的完整元信息容器(包含DbName、字段列表等) - DbBuild:数据库构建版本(如"Build 1.2") - Count:数据库内记录总数(纯数字) |
| 字段配置 | Field、FieldList | - Field:单个检索字段的配置(如PubMed的"Title"字段),包含: - IsDate(是否日期型:Y/N) - IsNumerical(是否数值型:Y/N) - IsTruncatable(是否支持截断检索:Y/N,如"cancer*"匹配"cancer""cancerous") - FieldList:多个Field的集合 |
| 链接配置 | Link、LinkList | - Link:数据库间的关联规则(如从"pubmed"链接到"protein") - LinkList:多个Link的集合 |
| 结果容器 | eInfoResult、DbList | - eInfoResult:XML根标签(所有返回数据的顶层容器) - DbList:所有可用数据库的DbName集合(如你示例中"pubmed""protein"等) |
| 错误/警告 | ERROR、Warning | - 当eInfo请求出错(如无效参数)时返回ERROR,警告信息返回Warning |
2. XML示例:eInfo的实际返回结果
我们print出来的XML片段是调用eInfo查询"所有Entrez数据库列表" 的结果,结构非常清晰:
xml
<?xml version="1.0" encoding="UTF-8" ?>
<!DOCTYPE eInfoResult PUBLIC "-//NLM//DTD einfo 20190110//EN" "https://eutils.ncbi.nlm.nih.gov/eutils/dtd/20190110/einfo.dtd">
<eInfoResult> <!-- 根标签 -->
<DbList> <!-- 数据库列表容器 -->
<DbName>pubmed</DbName> <!-- 文献数据库 -->
<DbName>protein</DbName> <!-- 蛋白质序列数据库 -->
<DbName>nuccore</DbName> <!-- 核心核苷酸序列数据库 -->
<DbName>ipg</DbName> <!-- 同源蛋白组数据库 -->
<!-- ... 其他数据库 ... -->
<DbName>gtr</DbName> <!-- 遗传检测登记数据库 -->
</DbList>
</eInfoResult>- 开头的
<!DOCTYPE ...>声明:指定该XML遵循的DTD版本(20190110版),确保解析器按正确规则解读; - 内容仅包含
<DbList>:说明这次eInfo请求的目标是"获取所有数据库名称";若请求某单个数据库(如PubMed)的详细信息,XML会包含<DbInfo>(含字段列表、链接列表等)。
3. 实际用途:如何用这些信息?
无论是DTD还是XML结果,核心用途都是支撑Entrez系统的检索开发或使用,常见场景包括:
1. 开发层面:解析eInfo返回数据
如果我们需要编写代码(如Python、Java)调用Entrez eInfo工具(通过NCBI的E-utilities API),DTD是"数据解析指南":
- 例如,用Python的
xml.etree.ElementTree解析XML时,可根据DTD确定"要提取的标签层级"(如eInfoResult -> DbList -> DbName); - 避免解析错误:比如知道
TermCount是纯数字,可直接转为整数,无需额外判断格式。
2. 使用层面:了解数据库检索规则
即使不开发,通过eInfo的XML结果也能帮我们更高效使用Entrez(如PubMed):
- 查可用字段:比如调用eInfo查询
pubmed的<FieldList>,可知道PubMed支持"Title"(标题)、"Author"(作者)、"MeSH Terms"(医学主题词)等检索字段; - 查关联数据库:比如从
<LinkList>知道PubMed的文献可直接链接到"protein"(相关蛋白质)、"gene"(相关基因)数据库,方便跨库检索。
4. 将这个XML读入到一个Python对象里面去
现在回到我们的result,因为这是一个非常简单的XML格式数据
而XML格式数据我们前面说了,我们有很多种方法可以进行解析

最原始的方法,就是将XML输出当做文本文件,那就是字符串数据,我们可以使用字符串查找以及处理函数来分析数据。
当然,最正常的做法应该是将XML格式数据解析为字典之类的python对象,这样我们才好编程。
使用 Bio.Entrez 的解析器, 我们可以直接将这个XML读入到一个Python对象里面去。
至于解析,前面提过parse和read两个API接口,大家可以再回过头去仔细看看。
此处使用read:
plain
from Bio import Entrez
handle = Entrez.einfo()
record = Entrez.read(handle)可以发现,record现在是一个拥有确定键值对的字典数据:

我这里简单打印出来看一下:
这个键对应的值存储了上面XML文件里面包含的数据库名字的列表:
plain
for key, value in record.items():
print(f"{key}:\n{"\n".join(value)}\n")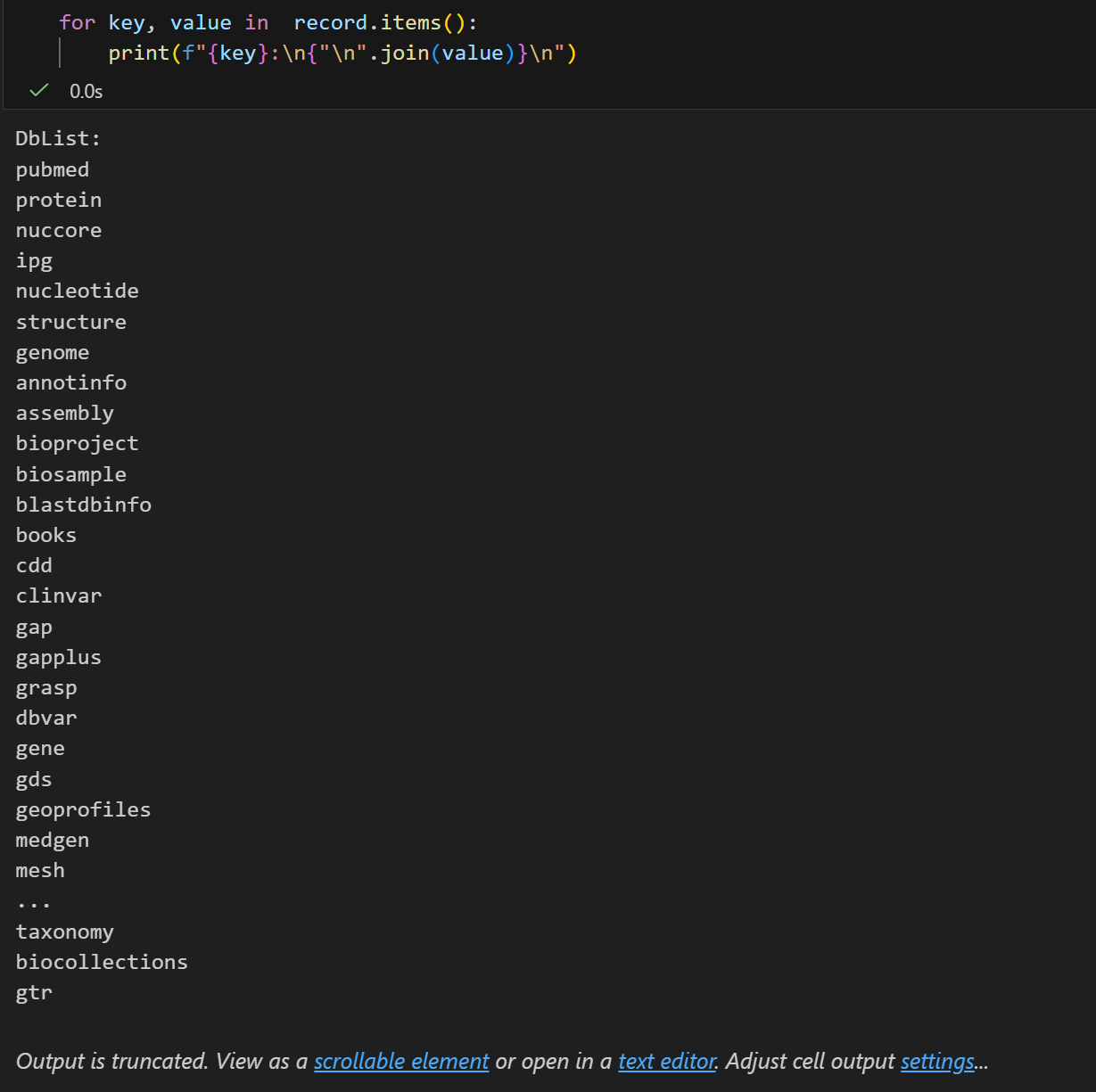

对于这些数据库,我们可以使用EInfo获得更多的信息:
比如说我们就看一下pubmed这个文献数据库,
plain
handle = Entrez.einfo(db="pubmed")
record = Entrez.read(handle)
还是只有一个键的字典,
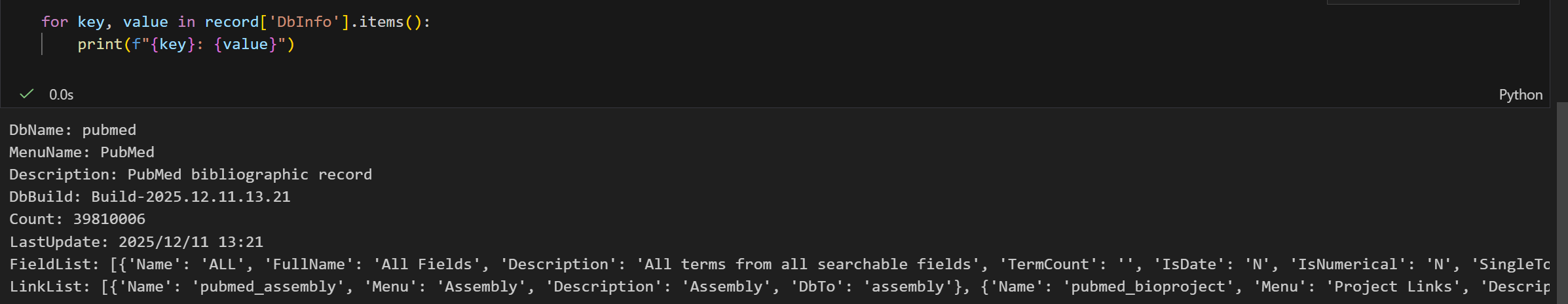
我们再仔细看看:
我们可以发现,lastupdate就是昨天,所以还是时效性上还是不用担心
主要是后面这两个字典list,
我们可以详细地展开看看
plain
for key, value in record['DbInfo'].items():
if key == 'FieldList':
print(f"{key}:")
for field in value:
for field_type, field_value in field.items():
print(f" {field_type}: {field_value}")
elif key == 'LinkList':
print(f"{key}:")
for link in value:
for link_type, link_value in link.items():
print(f" {link_type}: {link_value}")
else:
print(f"{key}: {value}")再次强调,该代码演示结果/数据时效日期为2025.12.12!
对于link链接部分,我们可以看到其实链接到的都是一些和组学相关的数据库,
plain
DbName: pubmed
MenuName: PubMed
Description: PubMed bibliographic record
DbBuild: Build-2025.12.11.13.21
Count: 39810006
LastUpdate: 2025/12/11 13:21
FieldList:
Name: ALL
FullName: All Fields
Description: All terms from all searchable fields
TermCount:
IsDate: N
IsNumerical: N
SingleToken: N
Hierarchy: N
IsHidden: N
Name: UID
FullName: UID
Description: Unique number assigned to publication
TermCount:
IsDate: N
IsNumerical: Y
SingleToken: Y
Hierarchy: N
IsHidden: Y
Name: FILT
FullName: Filter
Description: Limits the records
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: N
Name: TITL
FullName: Title
Description: Words in title of publication
TermCount:
IsDate: N
IsNumerical: N
SingleToken: N
Hierarchy: N
IsHidden: N
Name: MESH
FullName: MeSH Terms
Description: Medical Subject Headings assigned to publication
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: Y
IsHidden: N
Name: MAJR
FullName: MeSH Major Topic
Description: MeSH terms of major importance to publication
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: Y
IsHidden: N
Name: JOUR
FullName: Journal
Description: Journal abbreviation of publication
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: N
Name: AFFL
FullName: Affiliation
Description: Author's institutional affiliation and address
TermCount:
IsDate: N
IsNumerical: N
SingleToken: N
Hierarchy: N
IsHidden: N
Name: ECNO
FullName: EC/RN Number
Description: EC number for enzyme or CAS registry number
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: N
Name: SUBS
FullName: Supplementary Concept
Description: CAS chemical name or MEDLINE Substance Name
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: N
Name: PDAT
FullName: Date - Publication
Description: Date of publication
TermCount:
IsDate: Y
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: N
Name: EDAT
FullName: Date - Entry
Description: Date publication first accessible through Entrez
TermCount:
IsDate: Y
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: N
Name: VOL
FullName: Volume
Description: Volume number of publication
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: N
Name: PAGE
FullName: Pagination
Description: Page number(s) of publication
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: N
Name: PTYP
FullName: Publication Type
Description: Type of publication (e.g., review)
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: Y
IsHidden: N
Name: LANG
FullName: Language
Description: Language of publication
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: N
Name: ISS
FullName: Issue
Description: Issue number of publication
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: N
Name: SUBH
FullName: MeSH Subheading
Description: Additional specificity for MeSH term
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: Y
IsHidden: N
Name: SI
FullName: Secondary Source ID
Description: Cross-reference from publication to other databases
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: N
Name: MHDA
FullName: Date - MeSH
Description: Date publication was indexed with MeSH terms
TermCount:
IsDate: Y
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: N
Name: TIAB
FullName: Title/Abstract
Description: Free text associated with Abstract/Title
TermCount:
IsDate: N
IsNumerical: N
SingleToken: N
Hierarchy: N
IsHidden: N
Name: OTRM
FullName: Other Term
Description: Other terms associated with publication
TermCount:
IsDate: N
IsNumerical: N
SingleToken: N
Hierarchy: N
IsHidden: N
Name: COLN
FullName: Author - Corporate
Description: Corporate Author of publication
TermCount:
IsDate: N
IsNumerical: N
SingleToken: N
Hierarchy: N
IsHidden: N
Name: CNTY
FullName: Place of Publication
Description: Country of publication
TermCount:
IsDate: N
IsNumerical: N
SingleToken: N
Hierarchy: N
IsHidden: Y
Name: PAPX
FullName: Pharmacological Action
Description: MeSH pharmacological action pre-explosions
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: N
Name: GRNT
FullName: Grants and Funding
Description: NIH Grant Numbers
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: N
Name: MDAT
FullName: Date - Modification
Description: Date of last modification
TermCount:
IsDate: Y
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: N
Name: CDAT
FullName: Date - Completion
Description: Date of completion
TermCount:
IsDate: Y
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: N
Name: PID
FullName: Publisher ID
Description: Publisher ID
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: Y
Name: FAUT
FullName: Author - First
Description: First Author of publication
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: N
Name: FULL
FullName: Author
Description: Full Author Name(s) of publication
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: N
Name: FINV
FullName: Investigator
Description: Full name of investigator
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: N
Name: TT
FullName: Transliterated Title
Description: Words in transliterated title of publication
TermCount:
IsDate: N
IsNumerical: N
SingleToken: N
Hierarchy: N
IsHidden: N
Name: LAUT
FullName: Author - Last
Description: Last Author of publication
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: N
Name: PPDT
FullName: Print Publication Date
Description: Date of print publication
TermCount:
IsDate: Y
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: Y
Name: EPDT
FullName: Electronic Publication Date
Description: Date of Electronic publication
TermCount:
IsDate: Y
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: Y
Name: LID
FullName: Location ID
Description: ELocation ID
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: N
Name: CRDT
FullName: Date - Create
Description: Date publication first accessible through Entrez
TermCount:
IsDate: Y
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: N
Name: BOOK
FullName: Book
Description: ID of the book that contains the document
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: N
Name: ED
FullName: Editor
Description: Section's Editor
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: N
Name: ISBN
FullName: ISBN
Description: ISBN
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: N
Name: PUBN
FullName: Publisher
Description: Publisher's name
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: N
Name: AUCL
FullName: Author Cluster ID
Description: Author Cluster ID
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: Y
Name: EID
FullName: Extended PMID
Description: Extended PMID
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: Y
Name: AUID
FullName: Author - Identifier
Description: Author Identifier
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: N
Name: PS
FullName: Subject - Personal Name
Description: Personal Name as Subject
TermCount:
IsDate: N
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: N
Name: COIS
FullName: Conflict of Interest Statements
Description: Conflict of Interest Statements
TermCount:
IsDate: N
IsNumerical: N
SingleToken: N
Hierarchy: N
IsHidden: N
Name: WORD
FullName: Text Word
Description: Free text associated with publication
TermCount:
IsDate: N
IsNumerical: N
SingleToken: N
Hierarchy: N
IsHidden: N
Name: P1DAT
FullName: P1DAT
Description: Date publication first accessible through Solr
TermCount:
IsDate: Y
IsNumerical: N
SingleToken: Y
Hierarchy: N
IsHidden: Y
LinkList:
Name: pubmed_assembly
Menu: Assembly
Description: Assembly
DbTo: assembly
Name: pubmed_bioproject
Menu: Project Links
Description: Related Projects
DbTo: bioproject
Name: pubmed_biosample
Menu: BioSample Links
Description: BioSample links
DbTo: biosample
Name: pubmed_biosystems
Menu: BioSystem Links
Description: BioSystems
DbTo: biosystems
Name: pubmed_books_refs
Menu: Cited in Books
Description: PubMed links associated with Books
DbTo: books
Name: pubmed_cdd
Menu: Conserved Domain Links
Description: Link to related CDD entry
DbTo: cdd
Name: pubmed_clinvar
Menu: ClinVar
Description: Clinical variations associated with publication
DbTo: clinvar
Name: pubmed_clinvar_calculated
Menu: ClinVar (calculated)
Description: Clinical variations calculated to be associated with publication
DbTo: clinvar
Name: pubmed_dbvar
Menu: dbVar
Description: Link from PubMed to dbVar
DbTo: dbvar
Name: pubmed_gap
Menu: dbGaP Links
Description: Related dbGaP record
DbTo: gap
Name: pubmed_gds
Menu: GEO DataSet Links
Description: Related GEO DataSets
DbTo: gds
Name: pubmed_gene
Menu: Gene Links
Description: Link to related Genes
DbTo: gene
Name: pubmed_gene_bookrecords
Menu: Gene (from Bookshelf)
Description: Gene records in this citation
DbTo: gene
Name: pubmed_gene_citedinomim
Menu: Gene (OMIM) Links
Description: PubMed links to Gene derived from pubmed_omim_cited links
DbTo: gene
Name: pubmed_gene_pmc_nucleotide
Menu: Gene (nucleotide/PMC)
Description: Records in Gene identified from shared sequence and PMC links.
DbTo: gene
Name: pubmed_gene_rif
Menu: Gene (GeneRIF) Links
Description: Link to Gene for the GeneRIF subcategory
DbTo: gene
Name: pubmed_genome
Menu: Genome Links
Description: Published genome sequences
DbTo: genome
Name: pubmed_geoprofiles
Menu: GEO Profile Links
Description: GEO records associated with pubmed record
DbTo: geoprofiles
Name: pubmed_homologene
Menu: HomoloGene Links
Description: Related HomoloGene
DbTo: homologene
Name: pubmed_medgen
Menu: MedGen
Description: Related information in MedGen
DbTo: medgen
Name: pubmed_medgen_bookshelf_cited
Menu: MedGen (Bookshelf cited)
Description: Related records in MedGen based on citations in GeneReviews and Medical Genetics Summaries
DbTo: medgen
Name: pubmed_medgen_genereviews
Menu: MedGen (GeneReviews)
Description: Related MedGen records
DbTo: medgen
Name: pubmed_medgen_omim
Menu: MedGen (OMIM)
Description: Related information in MedGen (OMIM)
DbTo: medgen
Name: pubmed_nuccore
Menu: Nucleotide Links
Description: Published Nucleotide sequences
DbTo: nuccore
Name: pubmed_nuccore_refseq
Menu: Nucleotide (RefSeq) Links
Description: Link to Nucleotide RefSeqs
DbTo: nuccore
Name: pubmed_nuccore_weighted
Menu: Nucleotide (Weighted) Links
Description: Links to nuccore
DbTo: nuccore
Name: pubmed_omim_bookrecords
Menu: OMIM (from Bookshelf)
Description: OMIM records in this citation
DbTo: omim
Name: pubmed_omim_calculated
Menu: OMIM (calculated) Links
Description: OMIM (calculated) Links
DbTo: omim
Name: pubmed_omim_cited
Menu: OMIM (cited) Links
Description: OMIM (cited) Links
DbTo: omim
Name: pubmed_pcassay
Menu: PubChem BioAssay
Description: Related PubChem BioAssay
DbTo: pcassay
Name: pubmed_pccompound
Menu: PubChem Compound
Description: Related PubChem Compound
DbTo: pccompound
Name: pubmed_pccompound_mesh
Menu: PubChem Compound (MeSH Keyword)
Description: Related PubChem Compound via MeSH
DbTo: pccompound
Name: pubmed_pccompound_publisher
Menu: PubChem Compound (Publisher)
Description: Publisher deposited structures linked to PubChem Compound
DbTo: pccompound
Name: pubmed_pcsubstance
Menu: PubChem Substance Links
Description: Related PubChem Substance
DbTo: pcsubstance
Name: pubmed_pcsubstance_bookrecords
Menu: PubChem Substance (from Bookshelf)
Description: Structures in the PubChem Substance database in this citation
DbTo: pcsubstance
Name: pubmed_pcsubstance_publisher
Menu: PubChem Substance (Publisher)
Description: PubChem Substances supplied by publisher
DbTo: pcsubstance
Name: pubmed_pmc
Menu: PMC Links
Description: Free full text articles in PMC
DbTo: pmc
Name: pubmed_pmc_bookrecords
Menu: References in PMC for this Bookshelf citation
Description: Full text of articles in PubMed Central cited in this record
DbTo: pmc
Name: pubmed_pmc_embargo
Menu:
Description: Embargoed PMC article associated with PubMed
DbTo: pmc
Name: pubmed_pmc_local
Menu:
Description: Free full text articles in PMC
DbTo: pmc
Name: pubmed_pmc_refs
Menu: Cited in PMC
Description: PubMed links associated with PMC
DbTo: pmc
Name: pubmed_popset
Menu: PopSet Links
Description: Published population set
DbTo: popset
Name: pubmed_probe
Menu: Probe Links
Description: Related Probe entry
DbTo: probe
Name: pubmed_protein
Menu: Protein Links
Description: Published protein sequences
DbTo: protein
Name: pubmed_protein_refseq
Menu: Protein (RefSeq) Links
Description: Link to Protein RefSeqs
DbTo: protein
Name: pubmed_protein_weighted
Menu: Protein (Weighted) Links
Description: Links to protein
DbTo: protein
Name: pubmed_proteinclusters
Menu: Protein Cluster Links
Description: Related Protein Clusters
DbTo: proteinclusters
Name: pubmed_protfam
Menu: Protein Family Models
Description: Protein family models supported by a particular publication
DbTo: protfam
Name: pubmed_pubmed
Menu: Similar articles
Description: Similar PubMed articles, obtained by matching text and MeSH terms
DbTo: pubmed
Name: pubmed_pubmed_alsoviewed
Menu: Articles frequently viewed together
Description: Articles frequently viewed together
DbTo: pubmed
Name: pubmed_pubmed_bookrecords
Menu: References for this Bookshelf citation
Description: PubMed abstracts for articles cited in this record
DbTo: pubmed
Name: pubmed_pubmed_refs
Menu: References for PMC Articles
Description: References for this PMC Article
DbTo: pubmed
Name: pubmed_snp
Menu: SNP Links
Description: PubMed to SNP links
DbTo: snp
Name: pubmed_snp_cited
Menu: SNP (Cited)
Description: Related SNP (Cited) records
DbTo: snp
Name: pubmed_sra
Menu: SRA Links
Description: Links to Short Read Archive Experiments
DbTo: sra
Name: pubmed_structure
Menu: Structure Links
Description: Published 3D structures
DbTo: structure
Name: pubmed_taxonomy_entrez
Menu: Taxonomy via GenBank
Description: Related Taxonomy entry computed using other Entrez links
DbTo: taxonomy我们重点看一下这三个键值对:
plain
record['DbInfo']['Description'], record['DbInfo']['Count'], record['DbInfo']['LastUpdate']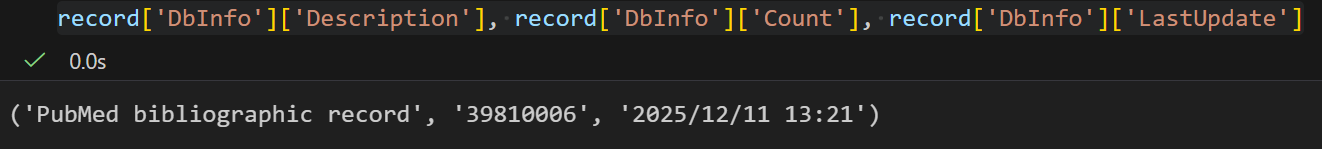
我们可以发现,截止到现在(2025.12.12),Pubmed中的文献记录也才4千万条左右,一个博士研究生可能4~5年左右也就大概看个1k篇左右的文献,总结就是文献是看不完的,就算筛选特定领域这个数目也是非常大的。
通过 record["DbInfo"].keys() 可以获取存储在这个记录里面的其他信息。这里面最有用的信息之一是一个ESearch可用的 搜索值列表:就是fieldlist
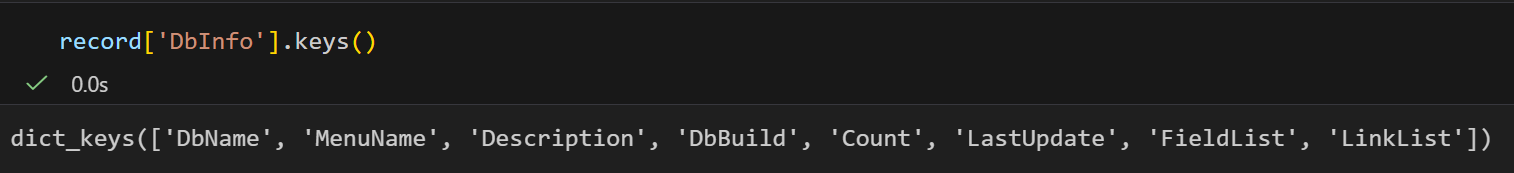
python
for field in record["DbInfo"]["FieldList"]:
print ("%(Name)s------%(FullName)s------%(Description)s" % field)都是一些对于pubmed检索来说比较重要的字段:
这是一个很长的列表,但是间接的告诉你在使用PubMed的时候,你可以通过 Jones[AUTH] 搜索作者,或者通过 Sanger[AFFL] 将作者范围限制在Sanger Centre。这个会非常方便,特别是在你对某个数据库不太熟悉的时候。
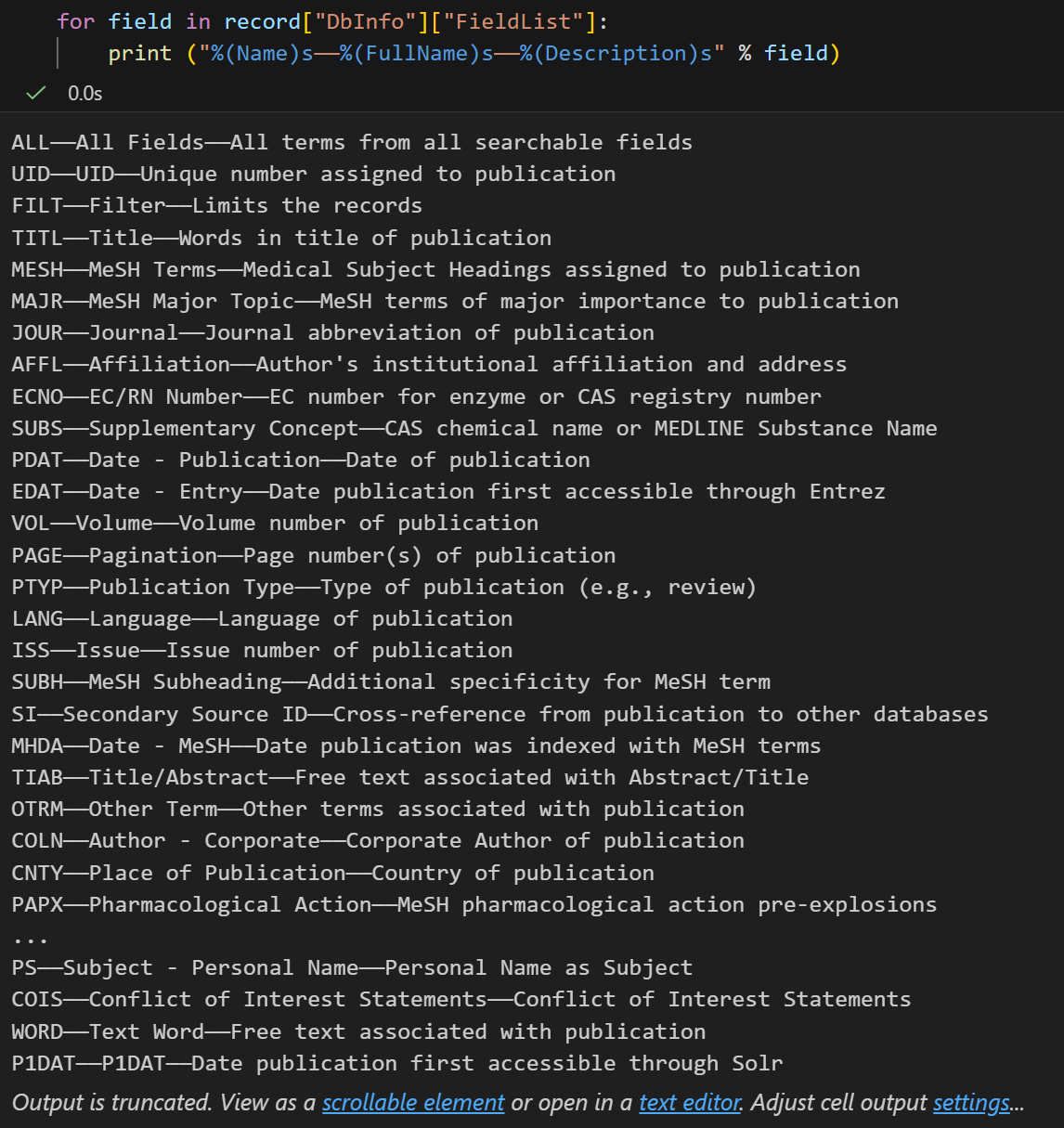
plain
ALL------All Fields------All terms from all searchable fields
UID------UID------Unique number assigned to publication
FILT------Filter------Limits the records
TITL------Title------Words in title of publication
MESH------MeSH Terms------Medical Subject Headings assigned to publication
MAJR------MeSH Major Topic------MeSH terms of major importance to publication
JOUR------Journal------Journal abbreviation of publication
AFFL------Affiliation------Author's institutional affiliation and address
ECNO------EC/RN Number------EC number for enzyme or CAS registry number
SUBS------Supplementary Concept------CAS chemical name or MEDLINE Substance Name
PDAT------Date - Publication------Date of publication
EDAT------Date - Entry------Date publication first accessible through Entrez
VOL------Volume------Volume number of publication
PAGE------Pagination------Page number(s) of publication
PTYP------Publication Type------Type of publication (e.g., review)
LANG------Language------Language of publication
ISS------Issue------Issue number of publication
SUBH------MeSH Subheading------Additional specificity for MeSH term
SI------Secondary Source ID------Cross-reference from publication to other databases
MHDA------Date - MeSH------Date publication was indexed with MeSH terms
TIAB------Title/Abstract------Free text associated with Abstract/Title
OTRM------Other Term------Other terms associated with publication
COLN------Author - Corporate------Corporate Author of publication
CNTY------Place of Publication------Country of publication
PAPX------Pharmacological Action------MeSH pharmacological action pre-explosions
GRNT------Grants and Funding------NIH Grant Numbers
MDAT------Date - Modification------Date of last modification
CDAT------Date - Completion------Date of completion
PID------Publisher ID------Publisher ID
FAUT------Author - First------First Author of publication
FULL------Author------Full Author Name(s) of publication
FINV------Investigator------Full name of investigator
TT------Transliterated Title------Words in transliterated title of publication
LAUT------Author - Last------Last Author of publication
PPDT------Print Publication Date------Date of print publication
EPDT------Electronic Publication Date------Date of Electronic publication
LID------Location ID------ELocation ID
CRDT------Date - Create------Date publication first accessible through Entrez
BOOK------Book------ID of the book that contains the document
ED------Editor------Section's Editor
ISBN------ISBN------ISBN
PUBN------Publisher------Publisher's name
AUCL------Author Cluster ID------Author Cluster ID
EID------Extended PMID------Extended PMID
AUID------Author - Identifier------Author Identifier
PS------Subject - Personal Name------Personal Name as Subject
COIS------Conflict of Interest Statements------Conflict of Interest Statements
WORD------Text Word------Free text associated with publication
P1DAT------P1DAT------Date publication first accessible through Solr5,ESearch: 搜索Entrez数据库
我们可以使用 Bio.Entrez.esearch() 来搜索任意的数据库。例如,我们在PubMed中搜索跟IDR相关的文献:
python
handle = Entrez.esearch(db="pubmed", term="idr")
record = Entrez.read(handle)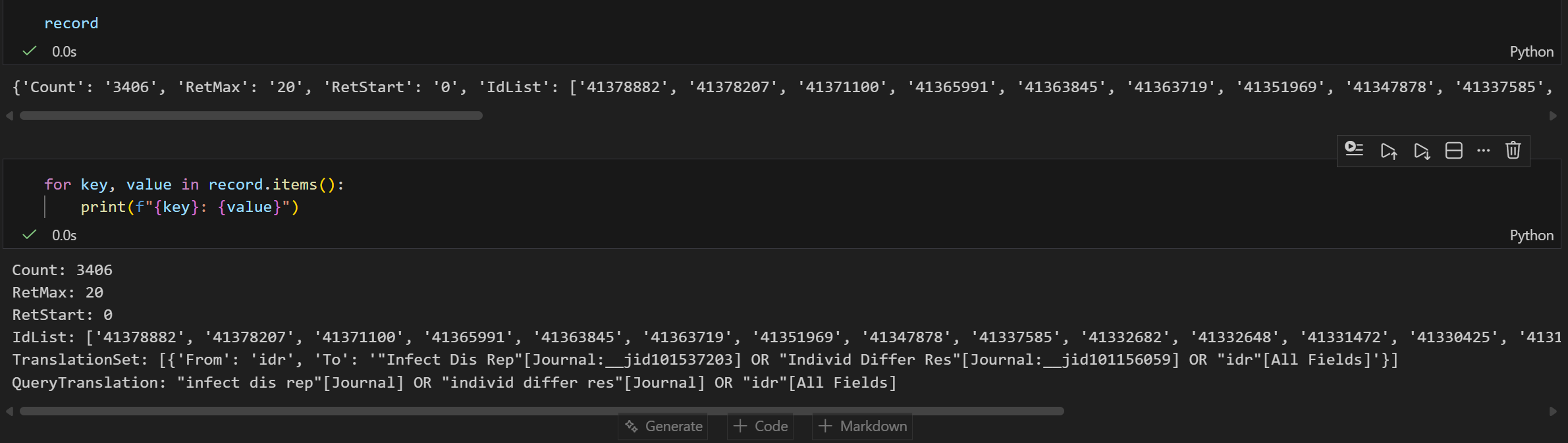
奇怪的是这里只返回20个pubmed ID,我一开始还乍一以为是idr这个term只能搜到20篇文献,但是猛的一看count都有3千多篇,而且我还设置了RetMax,所以只是限制返回的数目罢了,可以手动修改。
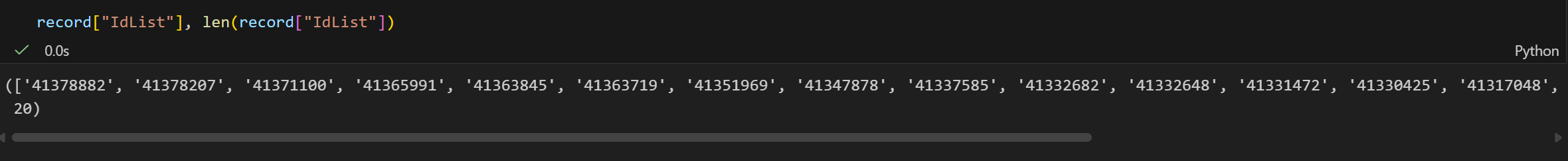
我们可以通过 EFetch来获取这些Pubmed IDs文献。
此处我们使用Efetch,一些参数设置细节参考NCBI官网https://www.ncbi.nlm.nih.gov/books/NBK25499/table/chapter4.T._valid_values_of__retmode_and/?report=objectonly

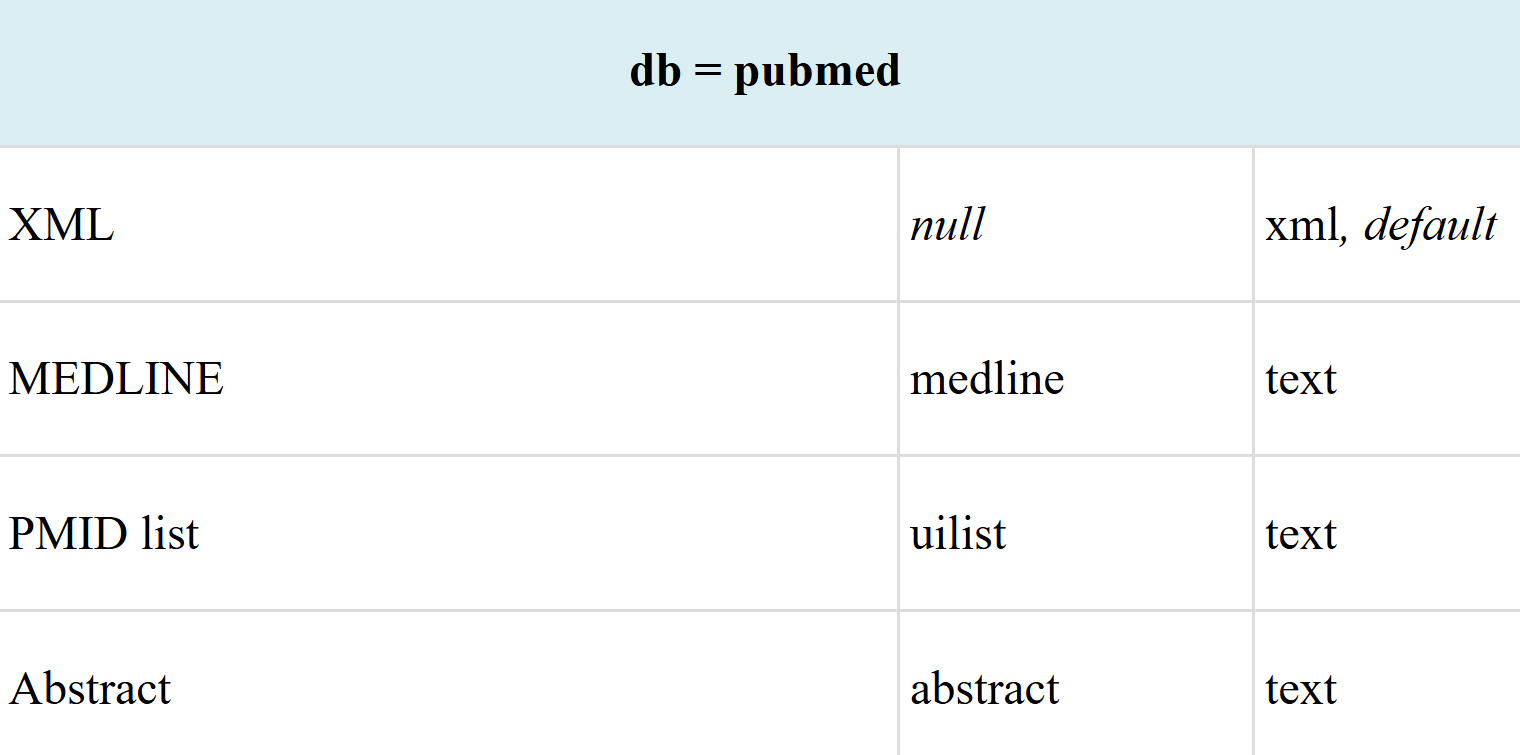
比如说我这里随便以41378882这个ID作为例子去解析,我们先返回Abstract类型看看:
python
handle = Entrez.efetch(db="pubmed", id="41378882", rettype="abstract", retmode="text")
print(handle.read()) # 注意因为上面retmode是text, 所以不需要handle再从XML解析了看起来返回的结果确实是挺像abstract的,而且文章也很新,
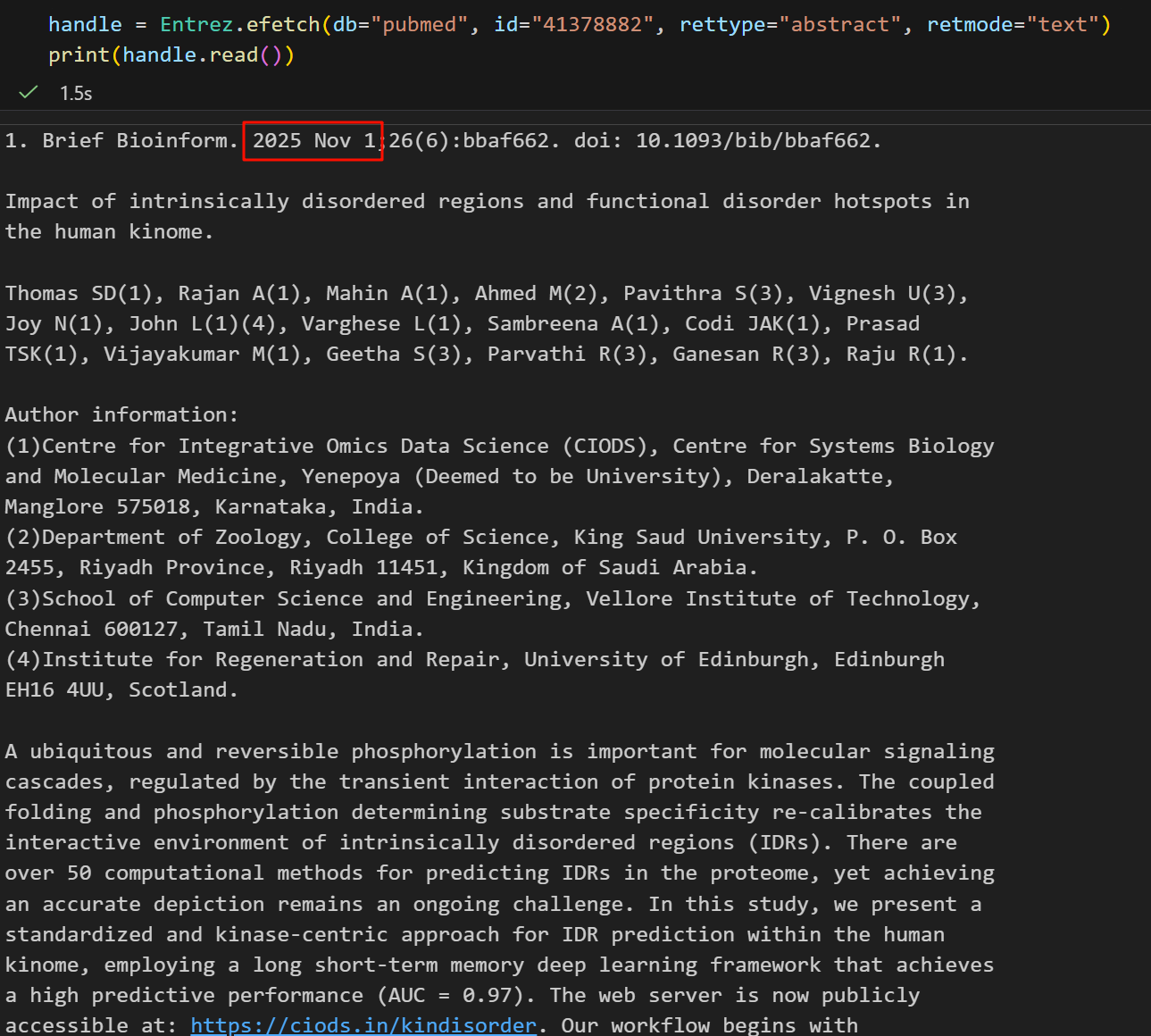
我把主要的内容拉下来放在下面
python
1. Brief Bioinform. 2025 Nov 1;26(6):bbaf662. doi: 10.1093/bib/bbaf662.
Impact of intrinsically disordered regions and functional disorder hotspots in
the human kinome.
Thomas SD(1), Rajan A(1), Mahin A(1), Ahmed M(2), Pavithra S(3), Vignesh U(3),
Joy N(1), John L(1)(4), Varghese L(1), Sambreena A(1), Codi JAK(1), Prasad
TSK(1), Vijayakumar M(1), Geetha S(3), Parvathi R(3), Ganesan R(3), Raju R(1).
Author information:
(1)Centre for Integrative Omics Data Science (CIODS), Centre for Systems Biology
and Molecular Medicine, Yenepoya (Deemed to be University), Deralakatte,
Manglore 575018, Karnataka, India.
(2)Department of Zoology, College of Science, King Saud University, P. O. Box
2455, Riyadh Province, Riyadh 11451, Kingdom of Saudi Arabia.
(3)School of Computer Science and Engineering, Vellore Institute of Technology,
Chennai 600127, Tamil Nadu, India.
(4)Institute for Regeneration and Repair, University of Edinburgh, Edinburgh
EH16 4UU, Scotland.
A ubiquitous and reversible phosphorylation is important for molecular signaling
cascades, regulated by the transient interaction of protein kinases. The coupled
folding and phosphorylation determining substrate specificity re-calibrates the
interactive environment of intrinsically disordered regions (IDRs). There are
over 50 computational methods for predicting IDRs in the proteome, yet achieving
an accurate depiction remains an ongoing challenge. In this study, we present a
standardized and kinase-centric approach for IDR prediction within the human
kinome, employing a long short-term memory deep learning framework that achieves
a high predictive performance (AUC = 0.97). The web server is now publicly
accessible at: https://ciods.in/kindisorder. Our workflow begins with
proteome-wide IDR prediction and proceeds with the categorization of short and
long IDR segments, followed by an in-depth analysis of their distribution
relative to the kinase domain regulatory core. We evaluated the conservation of
these IDRs across all 137 human kinase families, computing a trend-setting
conservation index to identify both conserved and variable disorder patterns.
Through this framework, we uncovered 1039 functional disorder region hotspots
that correlate with dynamic conformational shifts, phosphorylation sites,
functional motif enrichment, and mutation impact embedded within IDRs. To
further validate their regulatory significance, we conducted biophysical
profiling of conserved and variable IDRs. Finally, we developed a structural
integrity framework to link these IDRs to their influence on intrinsic signaling
cascades and substrate specificity. This study offers a comprehensive functional
characterization of IDRs in the human kinome, providing a valuable resource for
exploring kinase regulation and opportunities in drug repurposing.
© The Author(s) 2025. Published by Oxford University Press.
DOI: 10.1093/bib/bbaf662
PMID: 41378882 [Indexed for MEDLINE]其实我们肉眼就能够看出来,实际上的abstract有用的只有这么一小段
python
A ubiquitous and reversible phosphorylation is important for molecular signaling
cascades, regulated by the transient interaction of protein kinases. The coupled
folding and phosphorylation determining substrate specificity re-calibrates the
interactive environment of intrinsically disordered regions (IDRs). There are
over 50 computational methods for predicting IDRs in the proteome, yet achieving
an accurate depiction remains an ongoing challenge. In this study, we present a
standardized and kinase-centric approach for IDR prediction within the human
kinome, employing a long short-term memory deep learning framework that achieves
a high predictive performance (AUC = 0.97). The web server is now publicly
accessible at: https://ciods.in/kindisorder. Our workflow begins with
proteome-wide IDR prediction and proceeds with the categorization of short and
long IDR segments, followed by an in-depth analysis of their distribution
relative to the kinase domain regulatory core. We evaluated the conservation of
these IDRs across all 137 human kinase families, computing a trend-setting
conservation index to identify both conserved and variable disorder patterns.
Through this framework, we uncovered 1039 functional disorder region hotspots
that correlate with dynamic conformational shifts, phosphorylation sites,
functional motif enrichment, and mutation impact embedded within IDRs. To
further validate their regulatory significance, we conducted biophysical
profiling of conserved and variable IDRs. Finally, we developed a structural
integrity framework to link these IDRs to their influence on intrinsic signaling
cascades and substrate specificity. This study offers a comprehensive functional
characterization of IDRs in the human kinome, providing a valuable resource for
exploring kinase regulation and opportunities in drug repurposing.现在,我们正式在网页上检索看看,PMID: 41378882到底是不是这样的内容
注意,我们按照下面这种方式去检索
python
https://pubmed.ncbi.nlm.nih.gov/41378882/
# 也就是https://pubmed.ncbi.nlm.nih.gov/{PMID}/
经过我本人的比对check,
我们可以发现,其实是大差不差的,一些格式上不一致,但是无伤大雅,重要的一些文献中的数据都是一致的
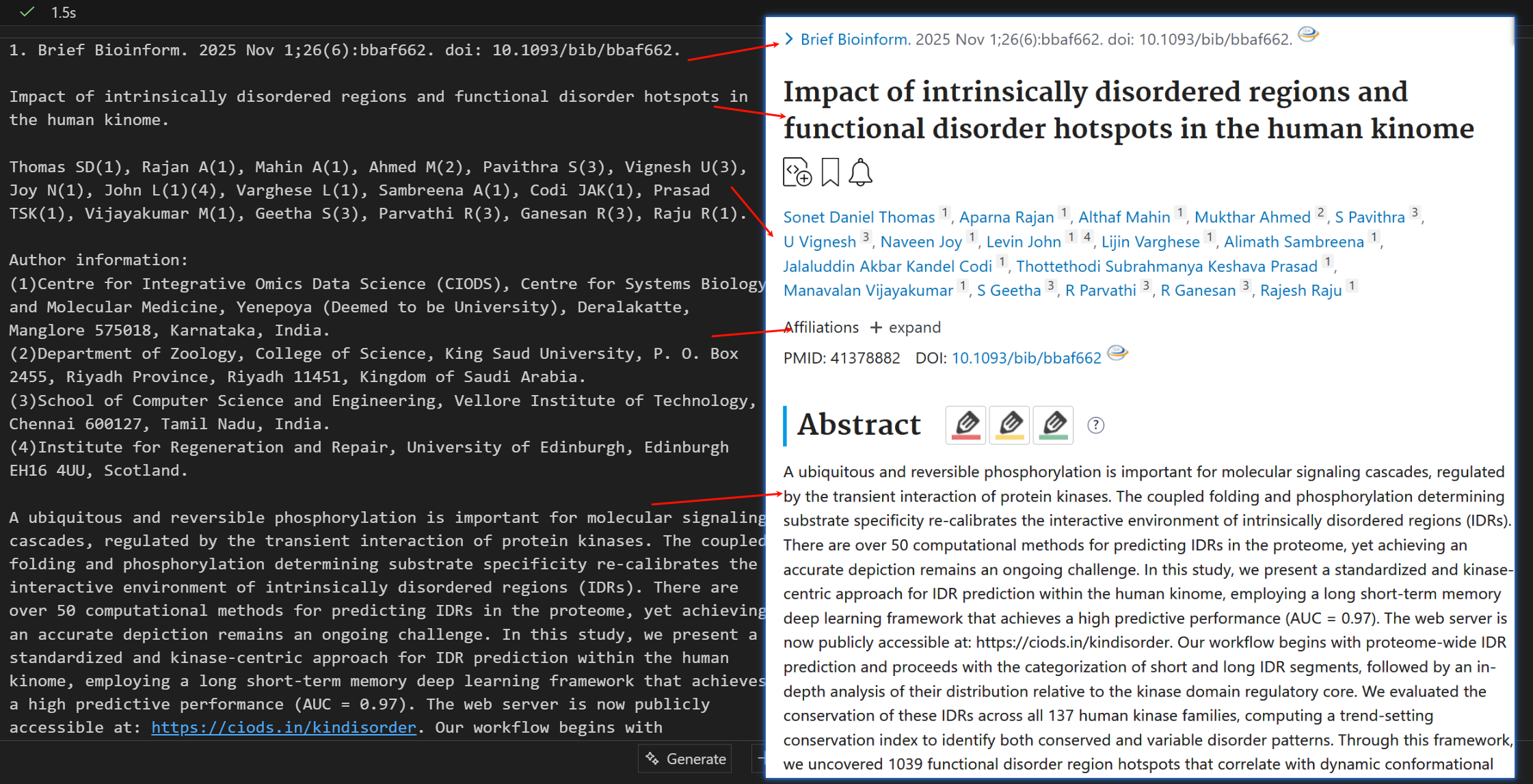
但是比较可惜的是,没有把这个keyword给扒取下来,
不然到时可以做文献集合的词云图,然后一些关键概念就可以收集起来,放在文献检索库里了。
总体来说还是不错的:因为是字符串对象,我们可以用上python强大的字符串处理工具,编写各种下游处理脚本,甚至可以再embedding化。
我们再来试一下Medline类型看看:
python
handle = Entrez.efetch(db="pubmed", id="41378882", rettype="medline", retmode="text")
print(handle.read())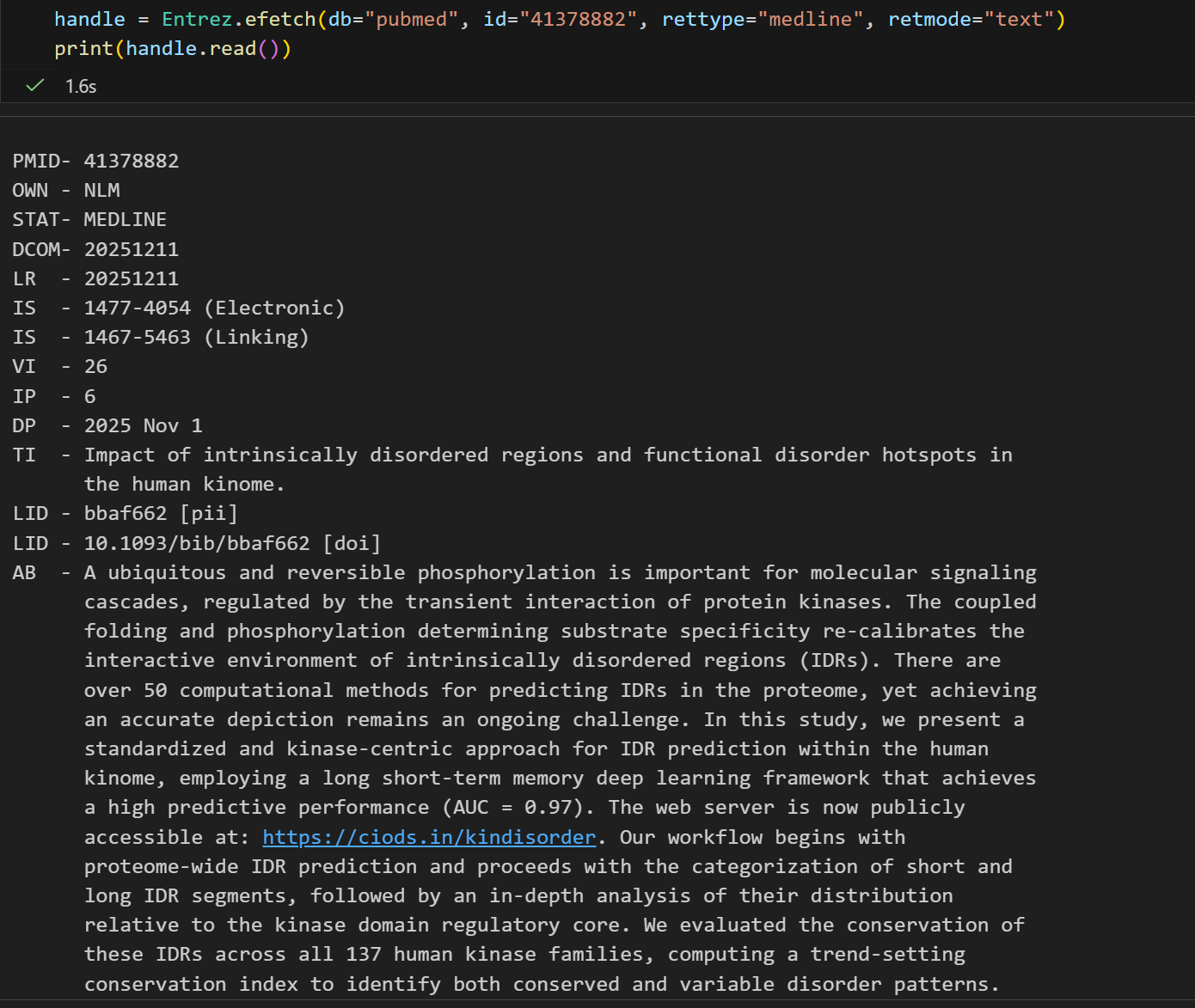
整体的记录格式会更简洁一点,
python
PMID- 41378882
OWN - NLM
STAT- MEDLINE
DCOM- 20251211
LR - 20251211
IS - 1477-4054 (Electronic)
IS - 1467-5463 (Linking)
VI - 26
IP - 6
DP - 2025 Nov 1
TI - Impact of intrinsically disordered regions and functional disorder hotspots in
the human kinome.
LID - bbaf662 [pii]
LID - 10.1093/bib/bbaf662 [doi]
AB - A ubiquitous and reversible phosphorylation is important for molecular signaling
cascades, regulated by the transient interaction of protein kinases. The coupled
folding and phosphorylation determining substrate specificity re-calibrates the
interactive environment of intrinsically disordered regions (IDRs). There are
over 50 computational methods for predicting IDRs in the proteome, yet achieving
an accurate depiction remains an ongoing challenge. In this study, we present a
standardized and kinase-centric approach for IDR prediction within the human
kinome, employing a long short-term memory deep learning framework that achieves
a high predictive performance (AUC = 0.97). The web server is now publicly
accessible at: https://ciods.in/kindisorder. Our workflow begins with
proteome-wide IDR prediction and proceeds with the categorization of short and
long IDR segments, followed by an in-depth analysis of their distribution
relative to the kinase domain regulatory core. We evaluated the conservation of
these IDRs across all 137 human kinase families, computing a trend-setting
conservation index to identify both conserved and variable disorder patterns.
Through this framework, we uncovered 1039 functional disorder region hotspots
that correlate with dynamic conformational shifts, phosphorylation sites,
functional motif enrichment, and mutation impact embedded within IDRs. To further
validate their regulatory significance, we conducted biophysical profiling of
conserved and variable IDRs. Finally, we developed a structural integrity
framework to link these IDRs to their influence on intrinsic signaling cascades
and substrate specificity. This study offers a comprehensive functional
characterization of IDRs in the human kinome, providing a valuable resource for
exploring kinase regulation and opportunities in drug repurposing.
CI - (c) The Author(s) 2025. Published by Oxford University Press.
FAU - Thomas, Sonet Daniel
AU - Thomas SD
AD - Centre for Integrative Omics Data Science (CIODS), Centre for Systems Biology and
Molecular Medicine, Yenepoya (Deemed to be University), Deralakatte, Manglore
575018, Karnataka, India.
FAU - Rajan, Aparna
AU - Rajan A
AD - Centre for Integrative Omics Data Science (CIODS), Centre for Systems Biology and
Molecular Medicine, Yenepoya (Deemed to be University), Deralakatte, Manglore
575018, Karnataka, India.
FAU - Mahin, Althaf
AU - Mahin A
AD - Centre for Integrative Omics Data Science (CIODS), Centre for Systems Biology and
Molecular Medicine, Yenepoya (Deemed to be University), Deralakatte, Manglore
575018, Karnataka, India.
FAU - Ahmed, Mukthar
AU - Ahmed M
AD - Department of Zoology, College of Science, King Saud University, P. O. Box 2455,
Riyadh Province, Riyadh 11451, Kingdom of Saudi Arabia.
FAU - Pavithra, S
AU - Pavithra S
AD - School of Computer Science and Engineering, Vellore Institute of Technology,
Chennai 600127, Tamil Nadu, India.
FAU - Vignesh, U
AU - Vignesh U
AD - School of Computer Science and Engineering, Vellore Institute of Technology,
Chennai 600127, Tamil Nadu, India.
FAU - Joy, Naveen
AU - Joy N
AD - Centre for Integrative Omics Data Science (CIODS), Centre for Systems Biology and
Molecular Medicine, Yenepoya (Deemed to be University), Deralakatte, Manglore
575018, Karnataka, India.
FAU - John, Levin
AU - John L
AD - Centre for Integrative Omics Data Science (CIODS), Centre for Systems Biology and
Molecular Medicine, Yenepoya (Deemed to be University), Deralakatte, Manglore
575018, Karnataka, India.
AD - Institute for Regeneration and Repair, University of Edinburgh, Edinburgh EH16
4UU, Scotland.
FAU - Varghese, Lijin
AU - Varghese L
AD - Centre for Integrative Omics Data Science (CIODS), Centre for Systems Biology and
Molecular Medicine, Yenepoya (Deemed to be University), Deralakatte, Manglore
575018, Karnataka, India.
FAU - Sambreena, Alimath
AU - Sambreena A
AD - Centre for Integrative Omics Data Science (CIODS), Centre for Systems Biology and
Molecular Medicine, Yenepoya (Deemed to be University), Deralakatte, Manglore
575018, Karnataka, India.
FAU - Codi, Jalaluddin Akbar Kandel
AU - Codi JAK
AD - Centre for Integrative Omics Data Science (CIODS), Centre for Systems Biology and
Molecular Medicine, Yenepoya (Deemed to be University), Deralakatte, Manglore
575018, Karnataka, India.
FAU - Prasad, Thottethodi Subrahmanya Keshava
AU - Prasad TSK
AD - Centre for Integrative Omics Data Science (CIODS), Centre for Systems Biology and
Molecular Medicine, Yenepoya (Deemed to be University), Deralakatte, Manglore
575018, Karnataka, India.
FAU - Vijayakumar, Manavalan
AU - Vijayakumar M
AD - Centre for Integrative Omics Data Science (CIODS), Centre for Systems Biology and
Molecular Medicine, Yenepoya (Deemed to be University), Deralakatte, Manglore
575018, Karnataka, India.
FAU - Geetha, S
AU - Geetha S
AD - School of Computer Science and Engineering, Vellore Institute of Technology,
Chennai 600127, Tamil Nadu, India.
FAU - Parvathi, R
AU - Parvathi R
AD - School of Computer Science and Engineering, Vellore Institute of Technology,
Chennai 600127, Tamil Nadu, India.
FAU - Ganesan, R
AU - Ganesan R
AD - School of Computer Science and Engineering, Vellore Institute of Technology,
Chennai 600127, Tamil Nadu, India.
FAU - Raju, Rajesh
AU - Raju R
AUID- ORCID: 0000-0003-2319-121X
AD - Centre for Integrative Omics Data Science (CIODS), Centre for Systems Biology and
Molecular Medicine, Yenepoya (Deemed to be University), Deralakatte, Manglore
575018, Karnataka, India.
LA - eng
PT - Journal Article
PL - England
TA - Brief Bioinform
JT - Briefings in bioinformatics
JID - 100912837
RN - 0 (Intrinsically Disordered Proteins)
RN - EC 2.7.- (Protein Kinases)
RN - 0 (Proteome)
SB - IM
MH - Humans
MH - *Intrinsically Disordered Proteins/chemistry/metabolism/genetics
MH - *Protein Kinases/chemistry/metabolism/genetics
MH - Phosphorylation
MH - Deep Learning
MH - Computational Biology/methods
MH - *Proteome/metabolism/chemistry
OTO - NOTNLM
OT - IDR conformation map
OT - functional disorder hotspots
OT - human kinome
OT - long short-term memory
EDAT- 2025/12/11 13:04
MHDA- 2025/12/11 13:05
CRDT- 2025/12/11 09:04
PHST- 2025/06/04 00:00 [received]
PHST- 2025/10/25 00:00 [revised]
PHST- 2025/11/18 00:00 [accepted]
PHST- 2025/12/11 13:05 [medline]
PHST- 2025/12/11 13:04 [pubmed]
PHST- 2025/12/11 09:04 [entrez]
AID - 8377154 [pii]
AID - 10.1093/bib/bbaf662 [doi]
PST - ppublish
SO - Brief Bioinform. 2025 Nov 1;26(6):bbaf662. doi: 10.1093/bib/bbaf662.还记得左边的这个field字段吗?是不是有点眼熟,其实和前面einfo中抓取pubmed的fieldlist中的元素内容有点像,
python
handle1 = Entrez.einfo(db="pubmed")
record1 = Entrez.read(handle1)
for field in record1["DbInfo"]['FieldList']:
print ("%(Name)s------%(FullName)s------%(Description)s" % field)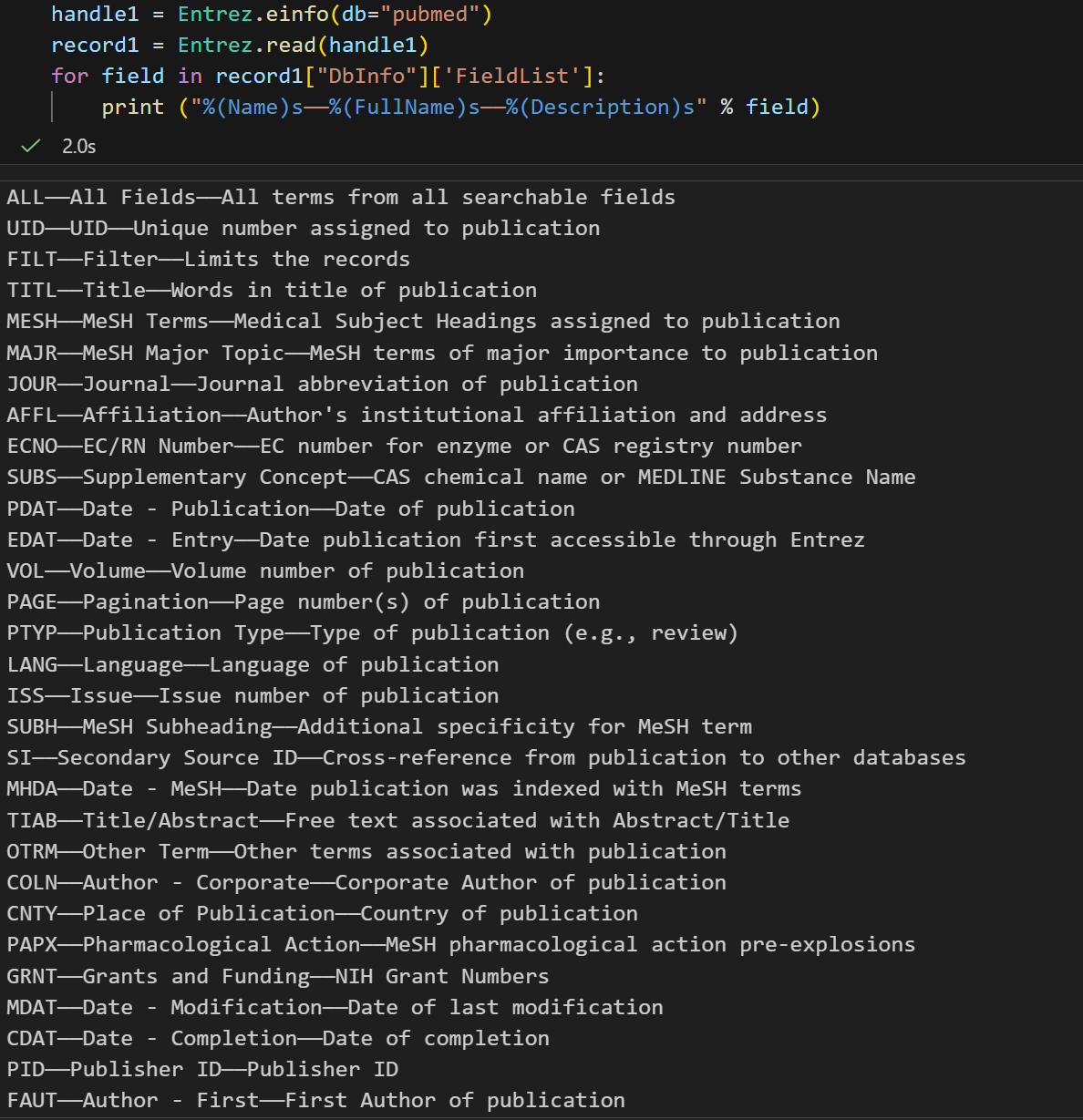
总之,medline类型返回的数据比abstract多,而且格式更加友好,
而且有些信息是显然对于我们从文献整理角度来看是非常有用的:
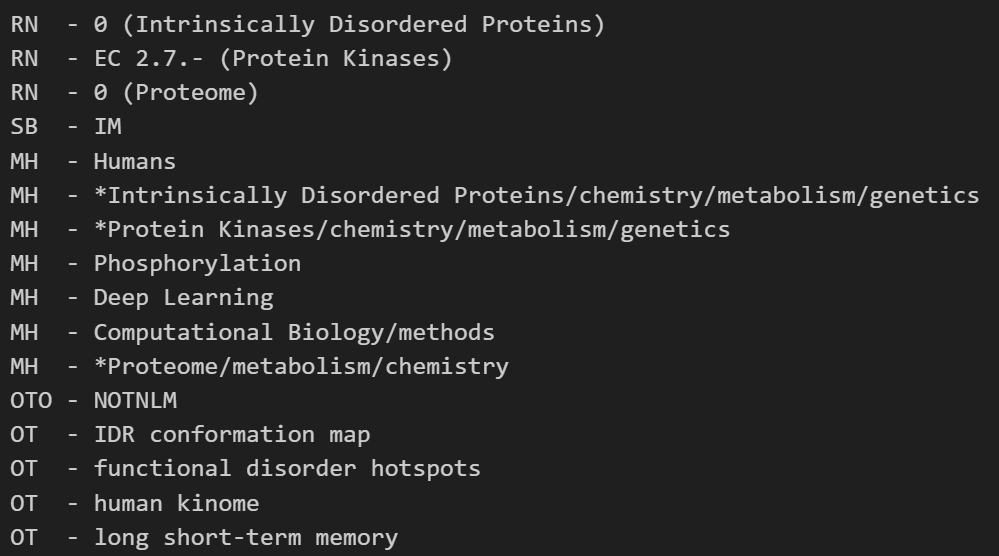
而且摘要我们可以直接从关键词上摘取:

再举一个例子,我们可以通过Esearch来搜索GenBank,此处以在人类中搜索BRCA1 gene为例,快速展示一下:
python
handle = Entrez.esearch(db="nucleotide", term="BRCA1[Gene] AND human[Organism]")
record = Entrez.read(handle)
handle.close()
record另外官网还有一个例子,参考:https://biopython-cn.readthedocs.io/zh-cn/latest/cn/chr09.html#sec-efetch
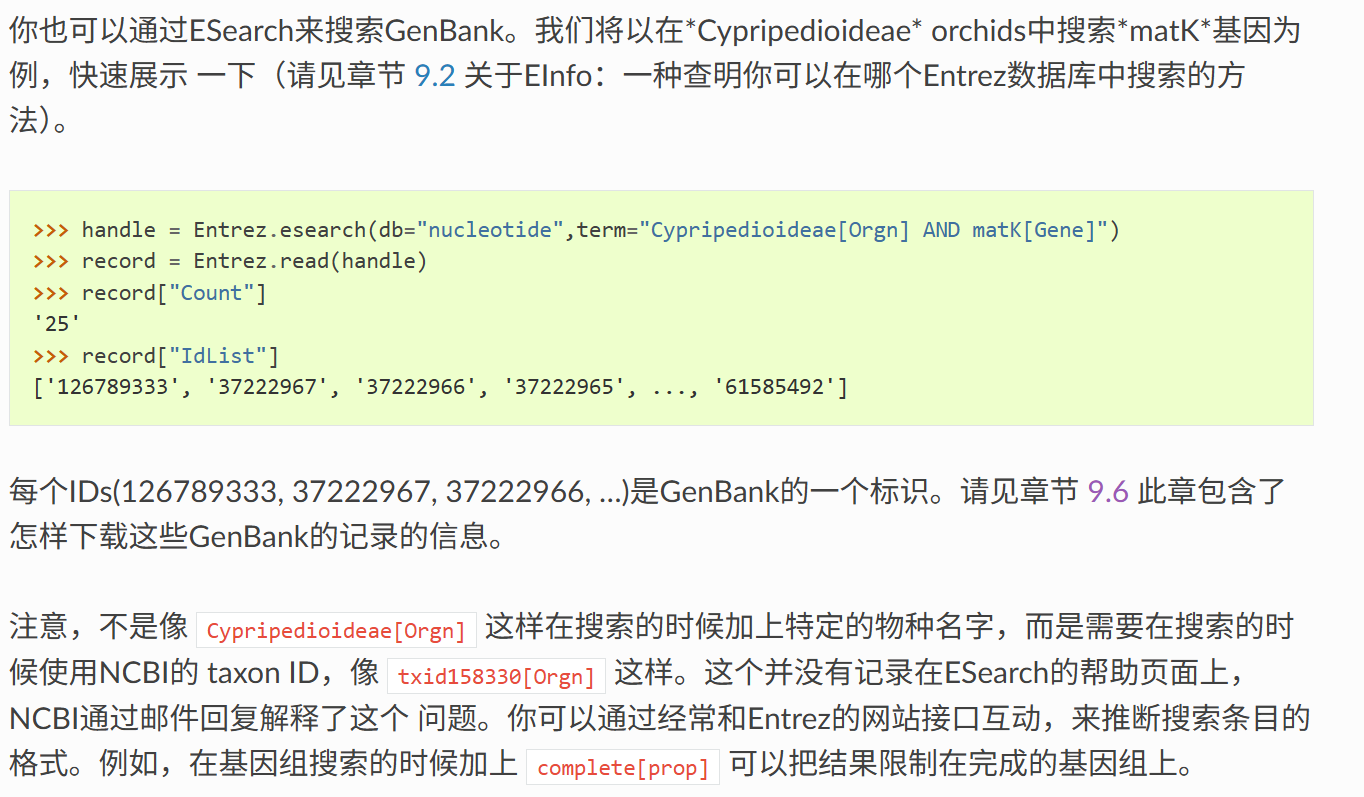
ESearch有很多有用的参数,具体可以参考Entrez官网使用说明:https://www.ncbi.nlm.nih.gov/books/NBK25499/#chapter4.ESearch
6,EPost: 上传identifiers的列表
具体细节参考官网:https://www.ncbi.nlm.nih.gov/books/NBK25499/#chapter4.EPost
EPost上传在后续搜索中将会用到的IDs的列表,通过 Bio.Entrez.epost() 函数可以在Biopython中实现。
就像前面我们在LibInspector模块结果解析中看到的那样,Epost本质上是将包含一组标识符(UIDs,如 PubMed 的 PMID、Protein 的 GI 号或 accession.version 等)的文件上传至 Entrez History 服务器,以便后续搜索策略(如 ESearch、EFetch、ELink 等操作)直接调用这组标识符,无需重复输入,提升效率。

参考官网:https://biopython-cn.readthedocs.io/zh-cn/latest/cn/chr09.html#sec-efetch
给出了一个简单的例子来看看EPost是如何工作的------上传了一些PubMed的IDs:

返回的XML包含了两个重要的字符串, QueryKey 和 WebEnv ,两个字符串一起确定了之前的历史记录。

我们可以使用其他的Entrez工具,例如EFetch,来提取这些值:
python
search_results = Entrez.read(Entrez.epost("pubmed", id=",".join(id_1ist)))
print(search_results)
webenv, query_key = search_results["WebEnv"], search_results["QueryKey"]
print(webenv, query_key)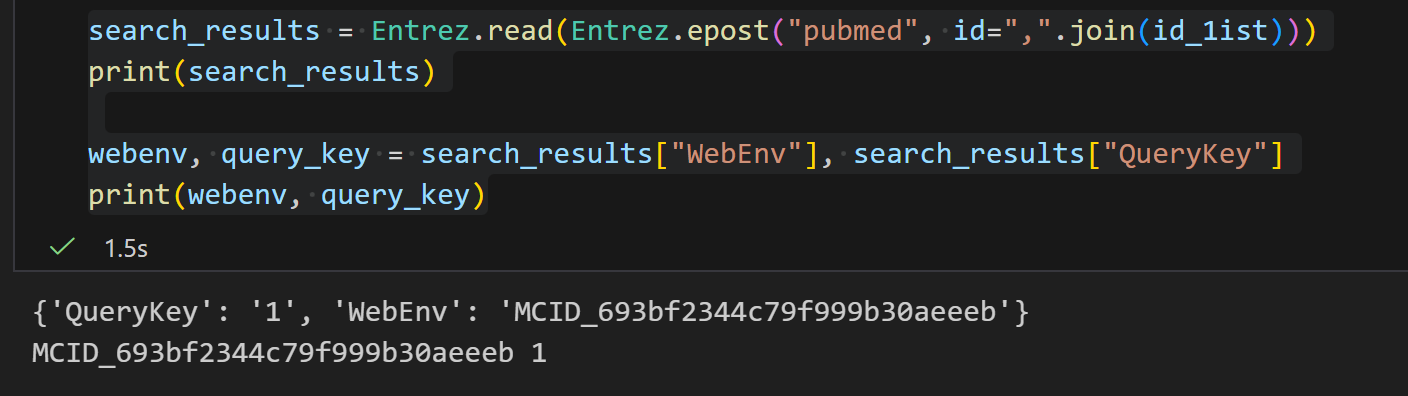
关于如何使用历史history的特性,我们会在后面进行介绍
7,ESummary: 通过主要的IDs来获取摘要
ESummary可以通过一个primary IDs来获取文章的摘要,详细细节可以参考官网:https://www.ncbi.nlm.nih.gov/books/NBK25499/#chapter4.ESummary。
在Biopython中,ESummary以 Bio.Entrez.esummary() 的形式出现。
我这里随便举一个例子,比如说获取ID为30367的pubmed文献相关的更多信息:
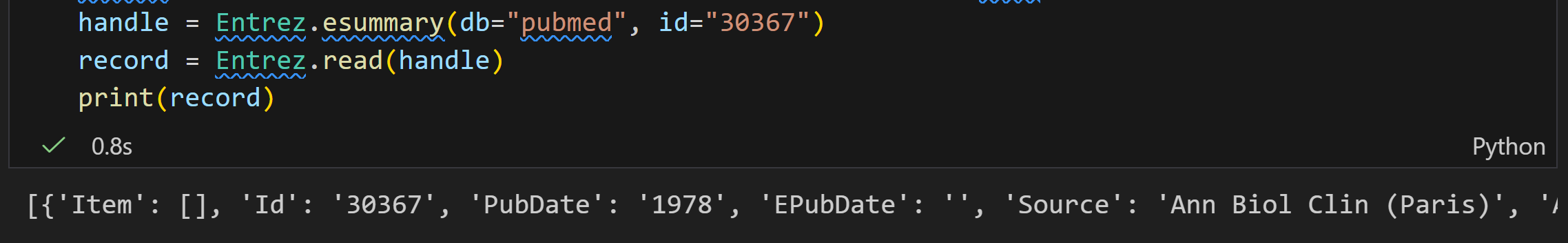
bash
record[0]['Id'], record[0]['Title'], record[0]['Source'], record[0]['PubDate']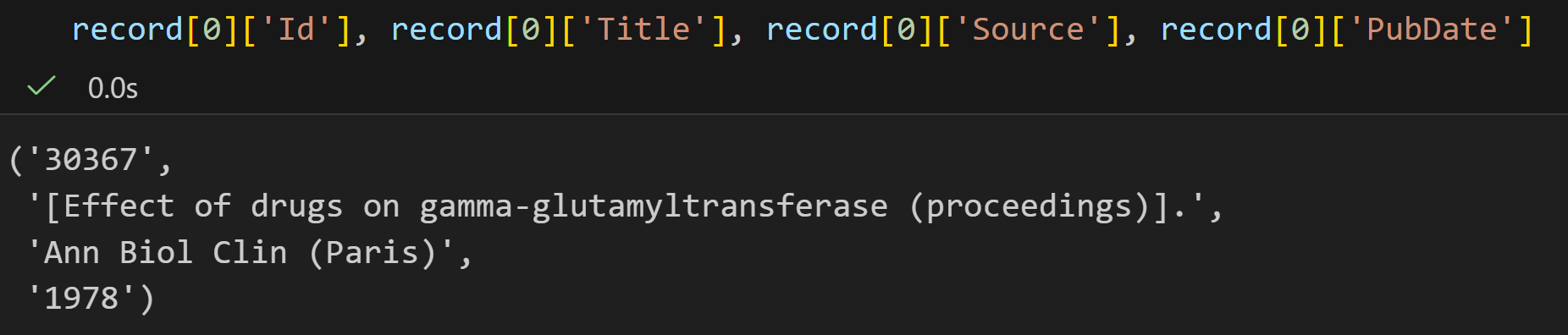
8,EFetch: 从Entrez下载更多的记录
当我们想要从Entrez中提取完整的记录的时候,我们可以使用EFetch。
具体细节,可以参考官网
https://www.ncbi.nlm.nih.gov/books/NBK25499/#chapter4.EFetch

其并不支持所有数据库,
具体支持的哪些数据库,可以参考:
https://www.ncbi.nlm.nih.gov/books/NBK25497/#chapter2.T._entrez_unique_identifiers_ui
我拉出来给大家看,就是下面的这些数据库是支持的:
因为我们这里只是尝试pubmed,是支持的
| Entrez Database | UID common name | E-utility Database Name |
|---|---|---|
| BioProject | BioProject ID | bioproject |
| BioSample | BioSample ID | biosample |
| Books | Book ID | books |
| Conserved Domains | PSSM-ID | cdd |
| dbGaP | dbGaP ID | gap |
| dbVar | dbVar ID | dbvar |
| Gene | Gene ID | gene |
| Genome | Genome ID | genome |
| GEO Datasets | GDS ID | gds |
| GEO Profiles | GEO ID | geoprofiles |
| HomoloGene | HomoloGene ID | homologene |
| MeSH | MeSH ID | mesh |
| NCBI C++ Toolkit | Toolkit ID | toolkit |
| NLM Catalog | NLM Catalog ID | nlmcatalog |
| Nucleotide | GI number | nuccore |
| PopSet | PopSet ID | popset |
| Probe | Probe ID | probe |
| Protein | GI number | protein |
| Protein Clusters | Protein Cluster ID | proteinclusters |
| PubChem BioAssay | AID | pcassay |
| PubChem Compound | CID | pccompound |
| PubChem Substance | SID | pcsubstance |
| PubMed | PMID | pubmed |
| PubMed Central | PMCID | pmc |
| SNP | rs number | snp |
| SRA | SRA ID | sra |
| Structure | MMDB-ID | structure |
| Taxonomy | TaxID | taxonomy |
NCBI大部分的数据库都支持多种不同的文件格式。当使用 Bio.Entrez.efetch() 从Entrez下载特定的某种格式的时候, 需要 rettype 和或者 retmode 这些可选的参数。对于不同数据库类型不同的搭配在下面的网页中有描述:
https://www.ncbi.nlm.nih.gov/books/NBK25499/#chapter4.EFetch
这个其实前面也提到过,现在我拉下来给大家看一下:
| Record Type | &rettype | &retmode |
|---|---|---|
| All Databases | ||
| Document summary | docsum | xml, default |
| List of UIDs in XML | uilist | xml |
| List of UIDs in plain text | uilist | text |
| db = bioproject | ||
| Full record XML | xml, default | xml, default |
| db = biosample | ||
| Full record XML | full, default | xml, default |
| Full record text | full, default | text |
| db = gds | ||
| Summary | summary, default | text, default |
| db = gene | ||
| text ASN.1 | null | asn.1, default |
| XML | null | xml |
| Gene table | gene_table | text |
| db = homologene | ||
| text ASN.1 | null | asn.1, default |
| XML | null | xml |
| Alignment scores | alignmentscores | text |
| FASTA | fasta | text |
| HomoloGene | homologene | text |
| db = mesh | ||
| Full record | full, default | text, default |
| db = nlmcatalog | ||
| Full record | null | text, default |
| XML | null | xml |
| db = nuccore, protein or popset | ||
| text ASN.1 | null | text, default |
| binary ASN.1 | null | asn.1 |
| Full record in XML | native | xml |
| Accession number(s) | acc | text |
| FASTA | fasta | text |
| TinySeq XML | fasta | xml |
| SeqID string | seqid | text |
| Additional options for db = nuccore or popset | ||
| GenBank flat file | gb | text |
| GBSeq XML | gb | xml |
| INSDSeq XML | gbc | xml |
| Additional option for db = nuccore and protein | ||
| Feature table | ft | text |
| Additional option for db = nuccore | ||
| GenBank flat file with full sequence (contigs) | gbwithparts | text |
| CDS nucleotide FASTA | fasta_cds_na | text |
| CDS protein FASTA | fasta_cds_aa | text |
| Additional options for db = protein | ||
| GenPept flat file | gp | text |
| GBSeq XML | gp | xml |
| INSDSeq XML | gpc | xml |
| Identical Protein XML | ipg | xml |
| db = pmc | ||
| XML | null | xml, default |
| MEDLINE | medline | text |
| db = pubmed | ||
| XML | null | xml*, default* |
| MEDLINE | medline | text |
| PMID list | uilist | text |
| Abstract | abstract | text |
| db = sequences | ||
| text ASN.1 | null | text, default |
| Accession number(s) | acc | text |
| FASTA | fasta | text |
| SeqID string | seqid | text |
| db = snp | ||
| text ASN.1 | null | asn.1, default |
| XML | null | xml |
| Flat file | flt | text |
| FASTA | fasta | text |
| RS Cluster report | rsr | text |
| SS Exemplar list | ssexemplar | text |
| Chromosome report | chr | text |
| Summary | docset | text |
| UID list | uilist | text or xml |
| db = sra | ||
| XML | full, default | xml, default |
| db = taxonomy | ||
| XML | null | xml, default |
| TaxID list | uilist | text or xml |
| db = clinvar | ||
| ClinVar Set | clinvarset | xml, default |
| UID list | uilist | text or xml |
| db = gtr | ||
| GTR Test Report | gtracc | xml, default |
一种常用的用法是下载FASTA或者GenBank/GenPept的文本格式 (接着可以使用 Bio.SeqIO 来解析),
此处还是以我们前面Esearch中检索的BRCA1 gene为例,
bash
handle = Entrez.esearch(db="nucleotide", term="BRCA1[Gene] AND human[Organism]")
record = Entrez.read(handle)
handle.close()
record因为要用到id,所以我们这里随便取1个:
就取第一个作为例子
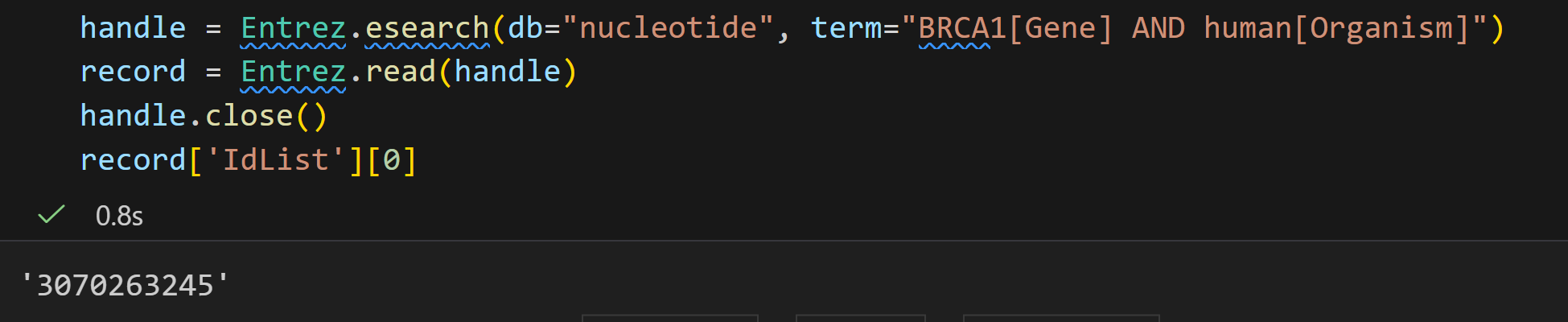
现在我们来看一下:
参数 rettype="gb" 和 retmode="text" 让我们下载的数据为GenBank格式。
python
handle = Entrez.efetch(db="nucleotide", id='3070263245', rettype="gb", retmode="text")
record = handle.read()
handle.close()
print(record)
完整的信息拉下来:
可以发现信息还是挺全的,时间也比较新,是今年2025年的,也给出了fasta序列。
bash
LOCUS PX436992 350 bp DNA linear PRI 07-OCT-2025
DEFINITION Homo sapiens isolate AB17 BRCA1 protein (BRCA1) gene, partial cds.
ACCESSION PX436992
VERSION PX436992.1
KEYWORDS .
SOURCE Homo sapiens (human)
ORGANISM Homo sapiens
Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi;
Mammalia; Eutheria; Euarchontoglires; Primates; Haplorrhini;
Catarrhini; Hominidae; Homo.
REFERENCE 1 (bases 1 to 350)
AUTHORS Abbas,A.F., Al-Shawi,A.A. and Ali,D.S.
TITLE Direct Submission
JOURNAL Submitted (27-SEP-2025) Department of Chemistry, University of
Basrah, Dur Al Thabat, Al Zubair, Basrah +964, Iraq
COMMENT ##Assembly-Data-START##
Sequencing Technology :: Sanger dideoxy sequencing
##Assembly-Data-END##
FEATURES Location/Qualifiers
source 1..350
/organism="Homo sapiens"
/mol_type="genomic DNA"
/isolate="AB17"
/db_xref="taxon:9606"
/chromosome="17"
/geo_loc_name="Iraq: Basrah"
/collection_date="2025-04-01"
/collected_by="Ali Falih Abbas"
gene <1..>350
/gene="BRCA1"
mRNA <30..>113
/gene="BRCA1"
/product="BRCA1 protein"
CDS <30..>113
/gene="BRCA1"
/note="breast cancer 1, early onset"
/codon_start=1
/product="BRCA1 protein"
/protein_id="YAW76721.1"
/translation="HDFEVRGDVVNGRNHQGPKRARESQDRK"
ORIGIN
1 cctgatgggt tgtgtttggt ttctttcagc atgattttga agtcagagga gatgtggtca
61 atggaagaaa ccaccaaggt ccaaagcgag caagagaatc ccaggacaga aaggtaaagc
121 tccctccctc aagttgacaa aaatctcacc ccaccactct gtattccact cccctttgca
181 gagatgggcc gcttcatttt gtaagactta ttacatacat acacagtgct agatactttc
241 acacaggttc ttttttcact cttccatccc aaccacataa ataagtattg tctctacttt
301 atgaatgata aaactaagag atttagagag gctgtgtatt ggggattccc
//需要注意的是直到2009年,Entrez EFetch API要求使用 "genbank" 作为返回类型,然而现在NCBI坚持使用官方的 "gb" 或 "gbwithparts" (或者针对蛋白的"gp") 返回类型。同样需要注意的是,直到2012年2月, Entrez EFetch API默认的返回格式为纯文本格式文件,现在默认的为XML格式。
作为另外的选择,我们也可以使用 rettype="fasta" 来获取Fasta格式的文件;
细节还是参考官网参数返回类型:https://www.ncbi.nlm.nih.gov/books/NBK25499/#chapter4.EFetch
也就是我上面拉下来的那一个长长的表格。
总之记住,可选的数据格式决定于我们要下载的数据库,我们要下载nucleotide,大概率就是返回gb;我们要下载pubmed,大概率就是返回text。
因为我们这里用的是BioPython,如果我们要获取记录的格式是Bio.SeqIO所接受的一种格式(详情参考biopython中的SeqIO模块,主要负责序列处理的输入输出,https://biopython-cn.readthedocs.io/zh-cn/latest/cn/chr05.html#chapter-bio-seqio)。
那么我们可以直接将其解析为一个 SeqRecord :
bash
from Bio import Entrez, SeqIO
Entrez.email = "Your email" # Always tell NCBI who you are
handle = Entrez.efetch(db="nucleotide", id='3070263245', rettype="gb", retmode="text")
record = SeqIO.read(handle, "genbank")
handle.close()
print(record)
完整的内容如下:
bash
ID: PX436992.1
Name: PX436992
Description: Homo sapiens isolate AB17 BRCA1 protein (BRCA1) gene, partial cds
Number of features: 4
/molecule_type=DNA
/topology=linear
/data_file_division=PRI
/date=07-OCT-2025
/accessions=['PX436992']
/sequence_version=1
/keywords=['']
/source=Homo sapiens (human)
/organism=Homo sapiens
/taxonomy=['Eukaryota', 'Metazoa', 'Chordata', 'Craniata', 'Vertebrata', 'Euteleostomi', 'Mammalia', 'Eutheria', 'Euarchontoglires', 'Primates', 'Haplorrhini', 'Catarrhini', 'Hominidae', 'Homo']
/references=[Reference(title='Direct Submission', ...)]
/structured_comment=defaultdict(<class 'dict'>, {'Assembly-Data': {'Sequencing Technology': 'Sanger dideoxy sequencing'}})
Seq('CCTGATGGGTTGTGTTTGGTTTCTTTCAGCATGATTTTGAAGTCAGAGGAGATG...CCC')需要注意的是,一种更加典型的用法是先把序列数据保存到一个本地文件,然后 使用 Bio.SeqIO 来解析。这样就避免了 在运行脚本的时候需要重复的下载同样的文件,并减轻NCBI服务器的负载。
我们还是用前面的BRCA1的第1个id为例:
bash
import os
from Bio import Entrez, SeqIO
Entrez.email = "your email" # Always tell NCBI who you are
filename = "gi_3070263245.gbk"
if not os.path.isfile(filename):
# Downloading...
net_handle = Entrez.efetch(db="nucleotide",id="3070263245",rettype="gb", retmode="text")
out_handle = open(filename, "w")
out_handle.write(net_handle.read())
out_handle.close()
net_handle.close()
print("Saved")
print("Parsing...")
record = SeqIO.read(filename, "genbank")
print(record)其实就是filename先存好了,后续访问会方便一点。
因为所有数据库都可以返回XML格式,并进一步解析为python中的对线,所以我们也可以试一下:
python
from Bio import Entrez
handle = Entrez.efetch(db="nucleotide", id="3070263245",retmode="xml")
record = Entrez.read(handle)
handle.close()
print(record)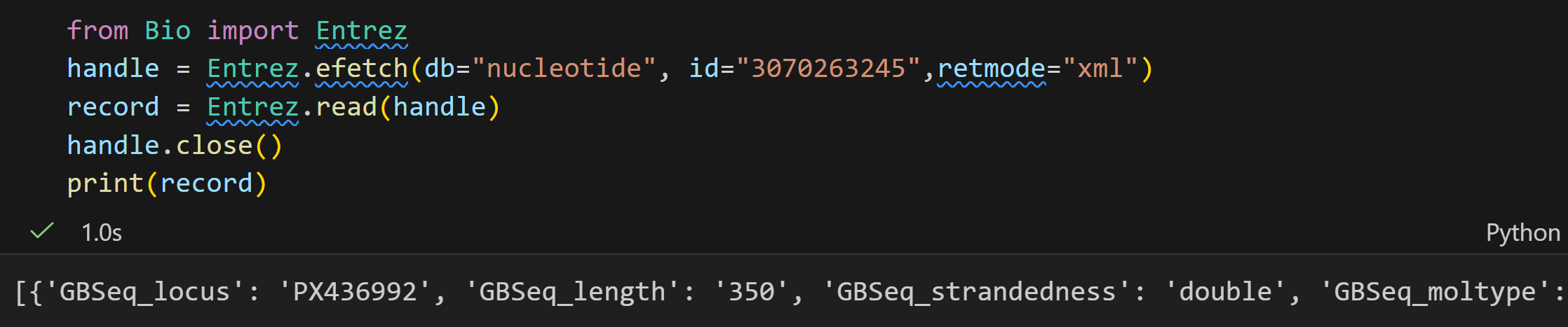
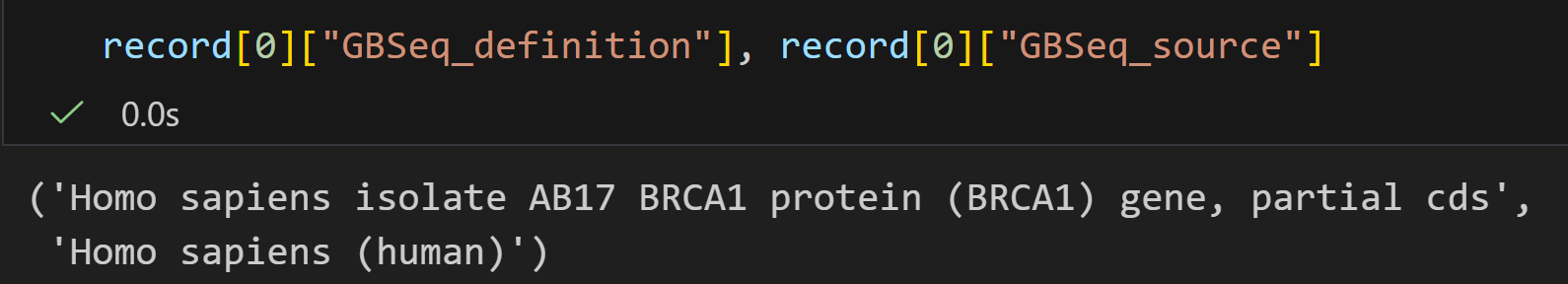
就像这样处理数据。例如解析其他数据库特异的文件格式(例如,PubMed中用到的 MEDLINE 格式)。
如果我们想使用 Bio.Entrez.esearch() 进行搜索,然后用 Bio.Entrez.efetch() 下载数据,那么我们需要用到 WebEnv的历史特性,就像上一节我提到的那样。
9,ELink: 在NCBI Entrez中搜索相关的条目
ELink,在Biopython中是 Bio.Entrez.elink() ,可以用来在NCBI Entrez数据库中寻找相关的条目。例如,我们以使用它在gene数据库中寻找核苷酸条目,或者其他很酷的事情。
此处,让我们用Elink来检索一篇PMID为40403066的文章,这篇文章是今年的一篇Science,
bash
pmid = "40403066"
record = Entrez.read(Entrez.elink(db="pubmed", id=pmid)) # 也可以写成handle再read解析
print(record)
变量 record 包含了一个Python列表,列出了已经搜索过的数据库。因为我们特指了一个PubMed ID来搜索,所以 record 只包含了一个条目。这个条目是一个字典变量,包含了我们需要寻找的条目的信息,以及能搜索到的所有相关 的内容:
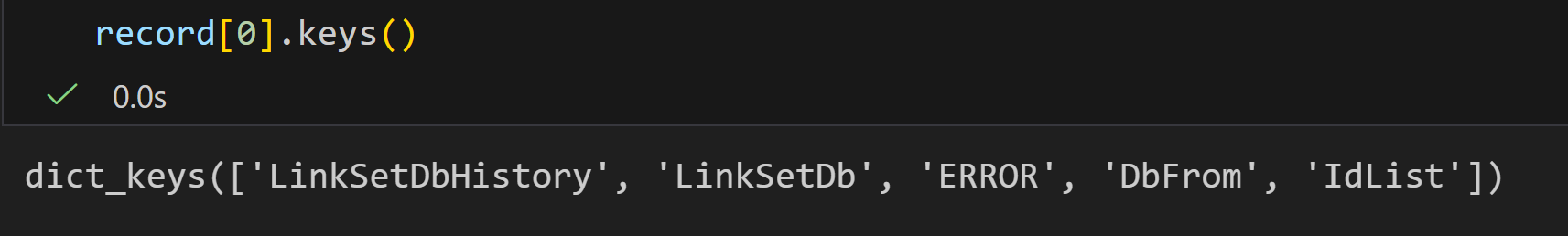
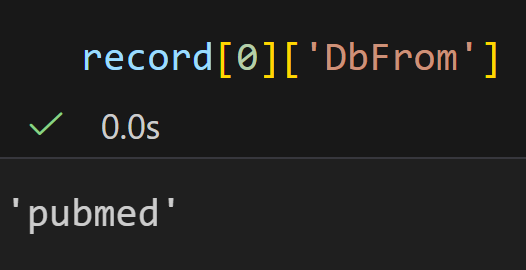
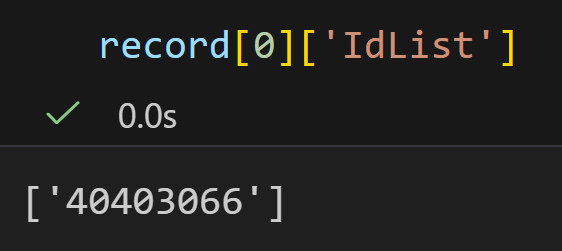
键 "LinkSetDb" 包含了搜索结果,将每个目标数据库保存为一个列表。在我们这个搜索中,我们只在PubMed数据库 中找到了结果(尽管已经被分到了不同的分类):
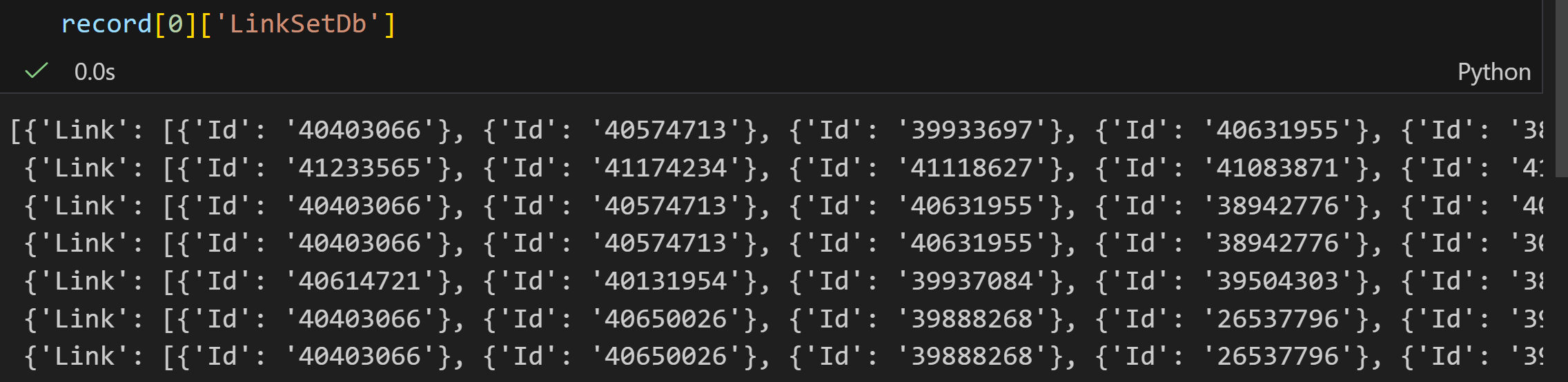
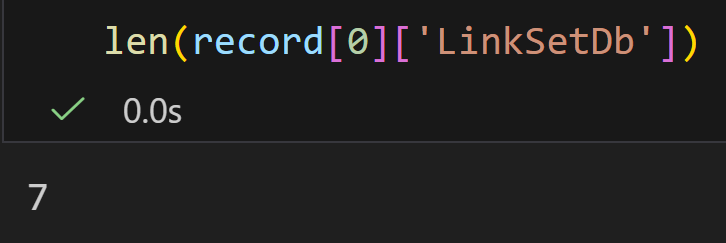
bash
for linksetdb in record[0]['LinkSetDb']:
print(f"Database: {linksetdb['DbTo']}, linknames: {linksetdb['LinkName']}, number of links: {len(linksetdb['Link'])}")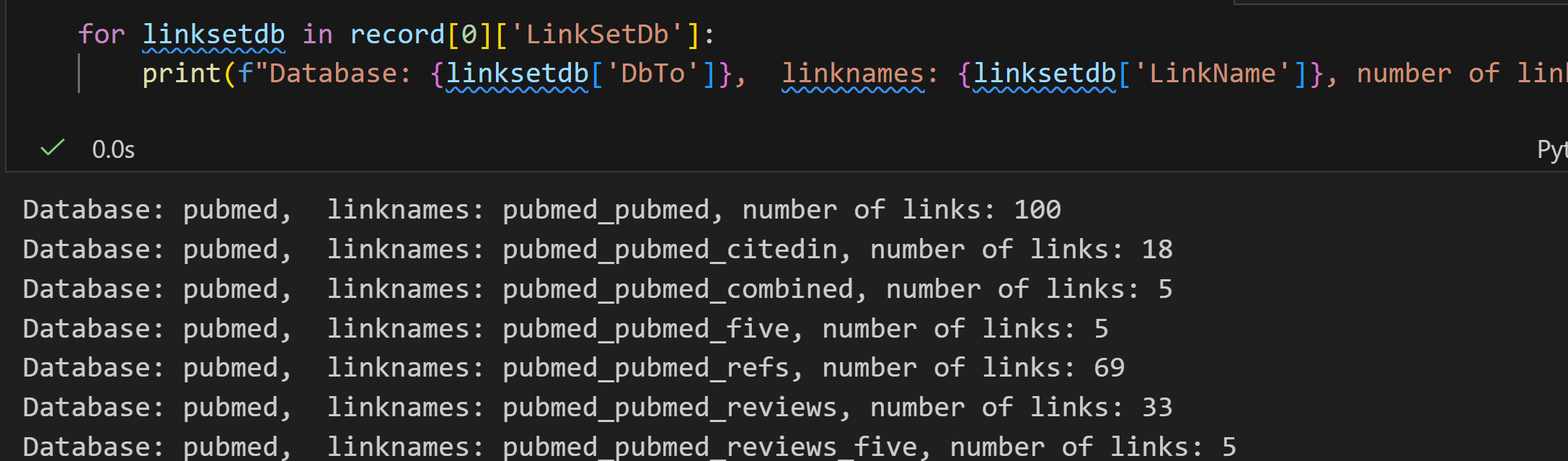
| LinkName | 含义(返回的 PMID 是什么) |
|---|---|
pubmed_pubmed |
相似/邻近文献(Related/Neighbors) :PubMed 的相似性算法给出的相关文献集合,常见上限 100 |
pubmed_pubmed_five |
上面"相似文献"的精选 5 条(用于界面快速展示) |
pubmed_pubmed_refs |
参考文献(References) :该 PMID 这篇文章引用了哪些 PubMed 文章(出链) |
pubmed_pubmed_citedin |
被引文献(Cited in) :有哪些 PubMed 文章引用了这篇文章(入链;取决于 NCBI 是否收录到引用关系) |
pubmed_pubmed_reviews |
综述(Reviews):与该文章相关联的"综述类"文章集合(通常可理解为围绕该主题/该文献的综述聚合) |
pubmed_pubmed_reviews_five |
上面"综述"的精选 5 条 |
pubmed_pubmed_combined |
综合集合:把多种关系(常见是相似/引用/被引等)做一个综合后的集合;有时也会是界面用的"合并展示"集合(所以你看到数量常较小/被截断) |
这其中有几个数据比较重要,首先是refs也就是引用的参考文献,这里的代码显示是69条,也就是说引用了69篇文献。
具体我们去https://pubmed.ncbi.nlm.nih.gov/40403066/ check一下:

整体显示有84篇引用文献,但是实际上我们看到有些引用文献是没有pubmed检索号的,所以可能refs没有统计进去,那么我亲自check了一下,发现一共有pubmed检索号的是70篇文献,
最后一篇索引号其实就是自己

所以,确实是69篇!
具体来说,这69篇索引逻辑是指收录在pubmed里的引用文献一共有69个,也就是说:
pubmed_pubmed_refs 返回的 "69 篇"指的是 NCBI 在 PubMed→PubMed 的引用链接里,能够成功匹配到对方记录(有 PMID 且被 PubMed 收录并建立了引用关系)的那一部分参考文献。其余参考文献并不一定"检索不到",而是不能通过这条 elink 的 PubMed 内部引用链接直接拿到 PMID。
常见原因:
- 参考文献本身不在 PubMed 收录范围内:书籍章节、会议摘要、学位论文、部分期刊/年代久远文献、非 MEDLINE 收录来源等 → 没有 PMID,自然不会出现在
pubmed_pubmed_refs。 - 引用条目类型不是传统论文:dataset、software、supporting data、专利、网页等 → 通常也没有 PMID。
- 有 PMID 但引用解析/匹配没建立链接:出版社元数据未提供、引用格式不规范、同名同刊歧义、NCBI 尚未完成引用关系抽取/更新 → 可能"在 PubMed 能搜到",但
elink不一定给出。 - 我们看到的"84 条参考文献"是文章正文参考文献总数;而
elink是 NCBI 的结构化链接集合,两者口径不同很正常。
如果我们想要拿到完整的参考文献列表,也就是包含那些没有PMID的条目,应该怎么做?
我们可以直接 efetch 取该 PMID 的 XML,然后再慢慢找哪些条目key是和reference相关的。
那么同样的,另外一个比较重要的就是他引了,我们想要看看这篇文献的基础上,其他人最新做了一些什么工作,那么就是看citation了。
从上面的结果中我们可以看到pubmed中的他引是18篇,
参考:https://pubmed.ncbi.nlm.nih.gov/?linkname=pubmed_pubmed_citedin&from_uid=40403066
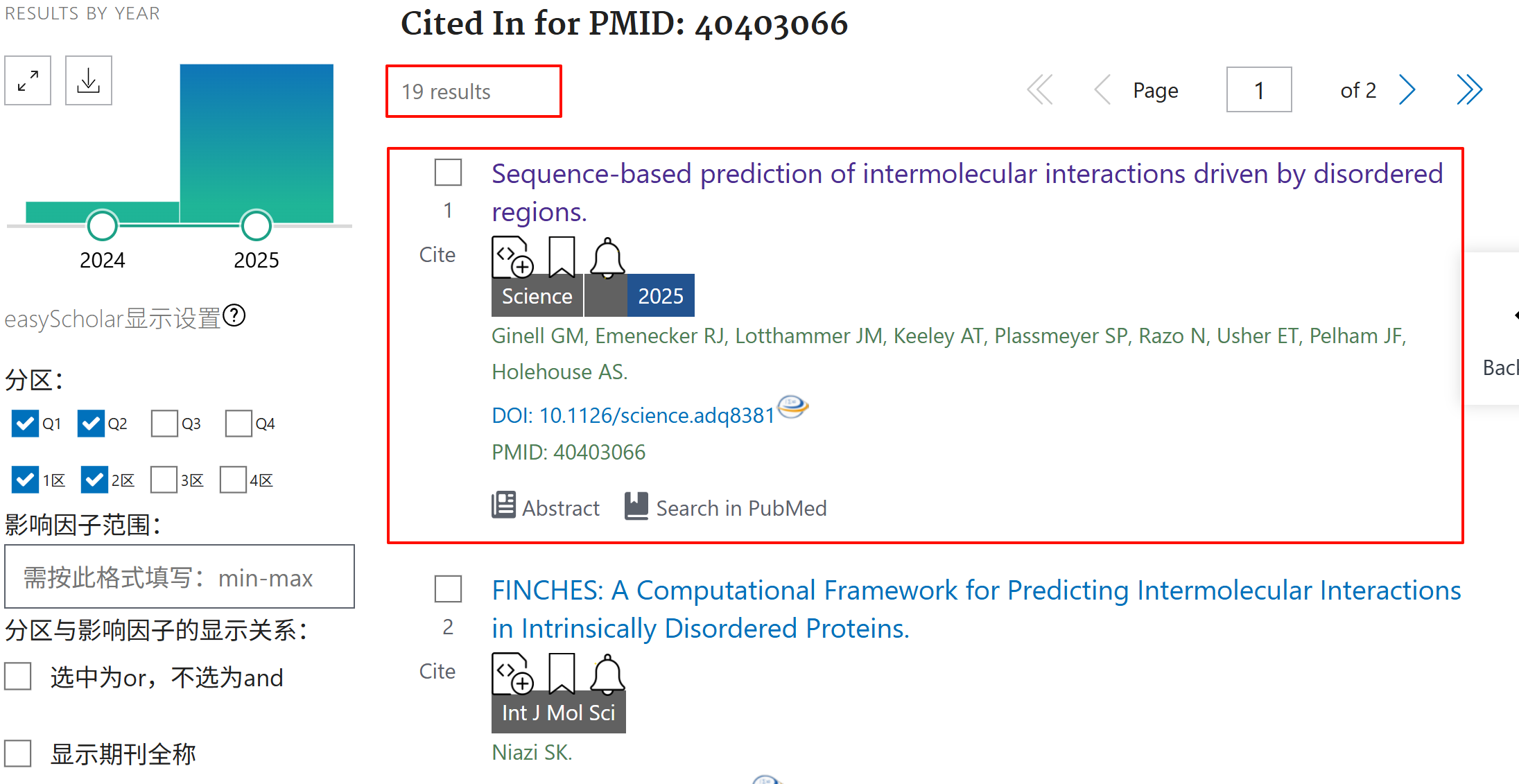
pubmed中显示是19篇,但是第一篇是自引不算,所以确实在pubmed中是18篇,当然其他非pubmed中,我们就不确定了,只能说我们获取的是一个子集数据。
但是至少,我们能够用Entrez Direct以及这个套壳的Biopython API,去获取我们感兴趣文献的大部分最新他引文献,这对于我们做文献调研非常有用。
现在回到我们前面讨论的LinkSetDb,实际的搜索结果被保存在键值为 "Link" 的字典下。
就像我们前面返回的结果所提示的那样,在标准搜索下,总共找到了100个条目, 现在让我们看看我们第一个的搜索结果:
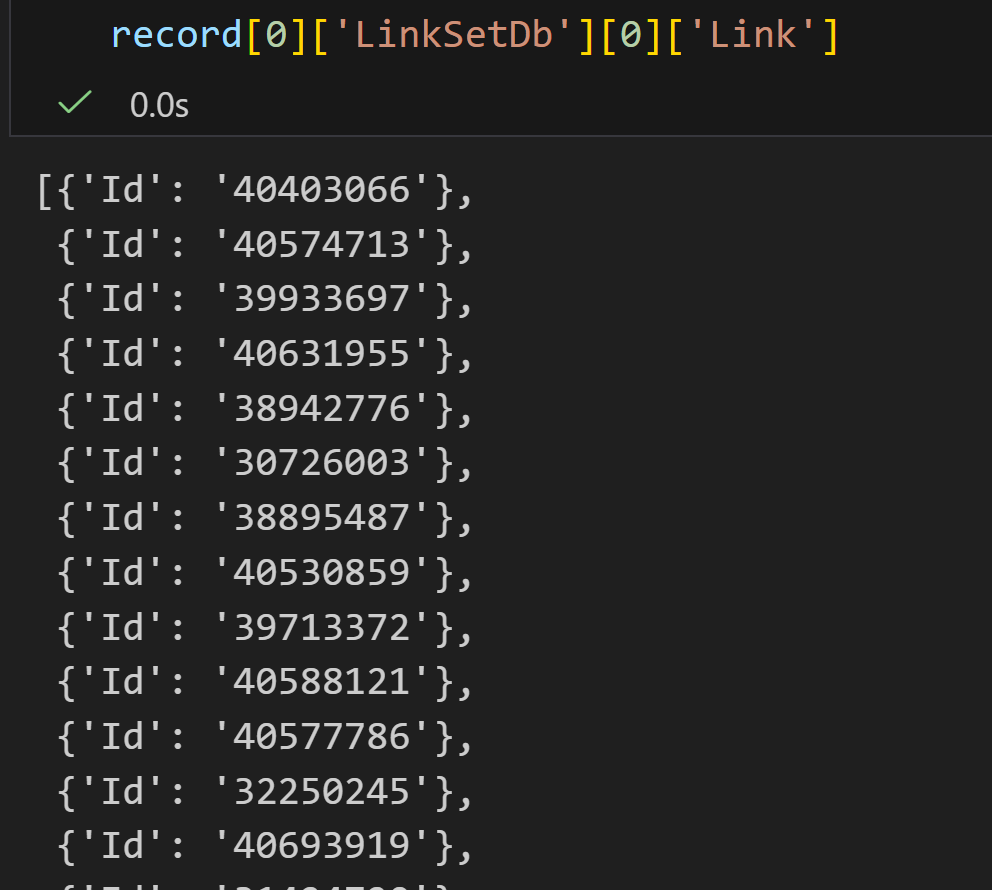
bash
record[0]['LinkSetDb'][0]['Link'][0]是不是很眼熟,没错,就是我们搜索的这篇文献本身!
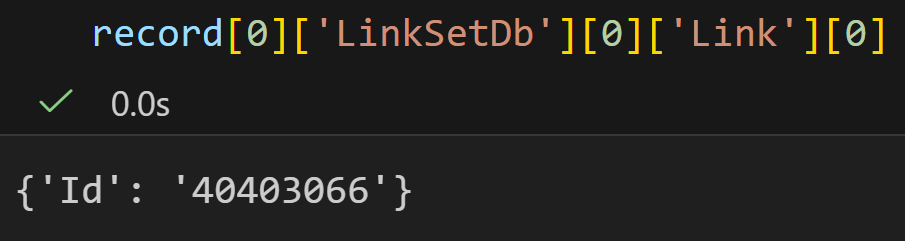
这个就是我们搜索的文章,从中并不能看到更多的结果,所以让我们来看看我们的第二个搜索结果:
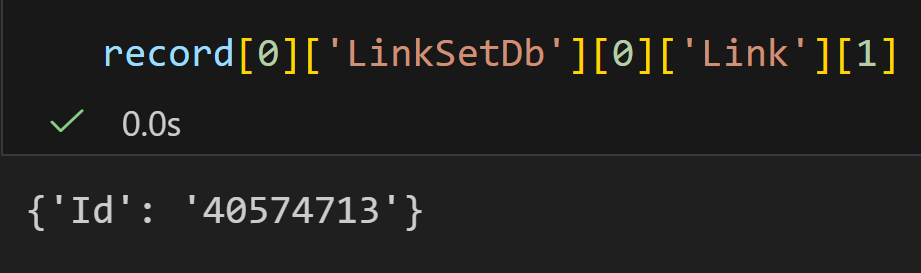
让我们来看看这篇PMID为40574713的文献到底是什么:

当然,我们可以通过一个循环来打印出所有的PubMed IDs:
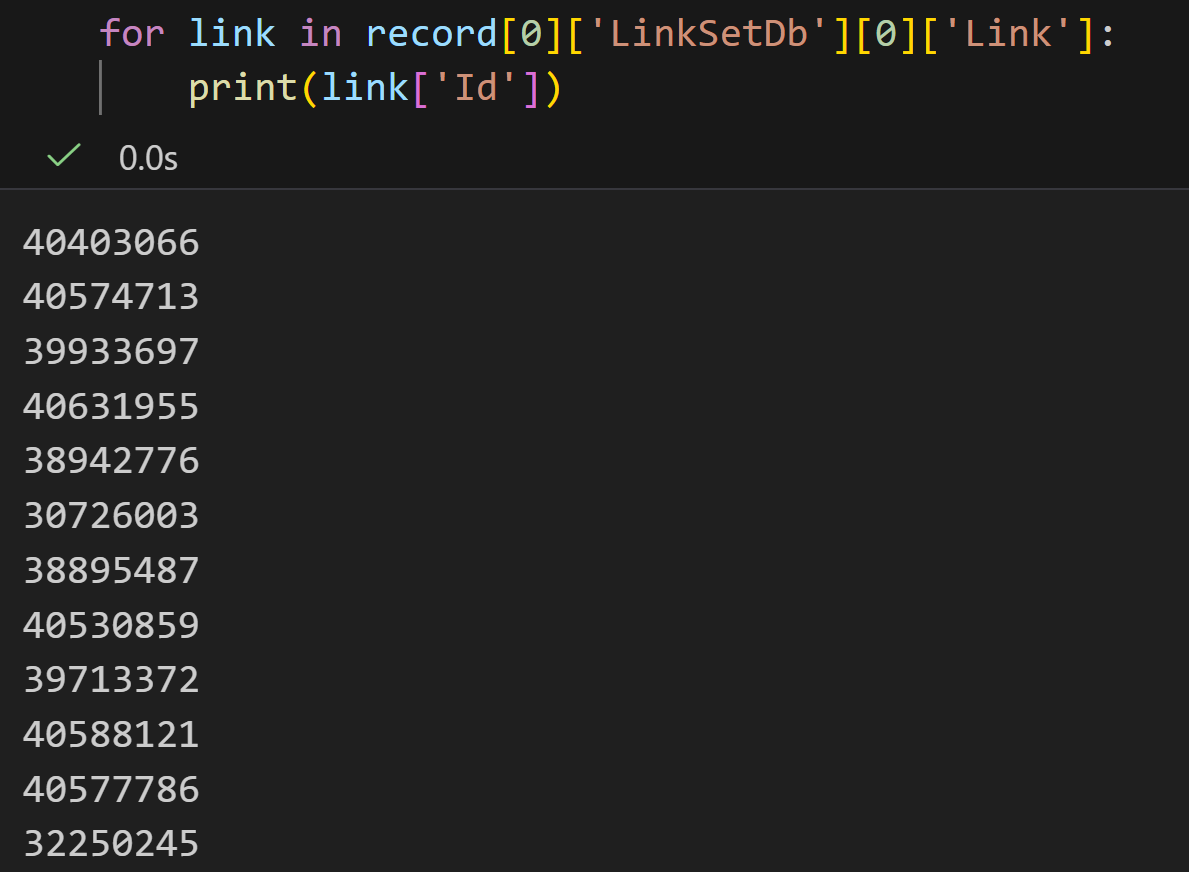
然后每一个link我们有了id,可以再用前面提到的Efetch。
但是对我个人而言,我对某篇文章是否被引用过更感兴趣,其实Elink也可以完成这个任务,至少对PubMed Central的杂志来说是这样的(但是引用全不全面,见我上面的那个例子)。
同样的,如果要深入细节,建议参考官网:https://www.ncbi.nlm.nih.gov/books/NBK25499/#chapter4.ELink
还有一个关于link name的整个子页面的详细参考,主要是描述了不同的数据库可以怎样交叉的索引:https://eutils.ncbi.nlm.nih.gov/corehtml/query/static/entrezlinks.html
这其中有家浙江这让我们
整体是
10,EGQuery: 全局搜索- 统计搜索的条目
EGQuery提供搜索字段在每个Entrez数据库中的数目。当我们只需要知道在每个数据库中能找到的条目的个数, 而不需要知道具体搜索结果的时候,这个非常的有用。
但是,就像我们前面用LibInspector看到的那样,这个工具NCBI官方已经不再支持了。
11,ESpell: 获得拼写建议
ESpell可以检索拼写建议。在这个例子中,我们使用 Bio.Entrez.espell() 来获得alphafold正确的拼写,
就像我们前面从LibInspector中看到的那个例子一样:
如果我一不小心在检索的时候敲了一个"Alphaffold"进去,
bash
handle = Entrez.espell(term="Alphaffold")
record = Entrez.read(handle)
print(record)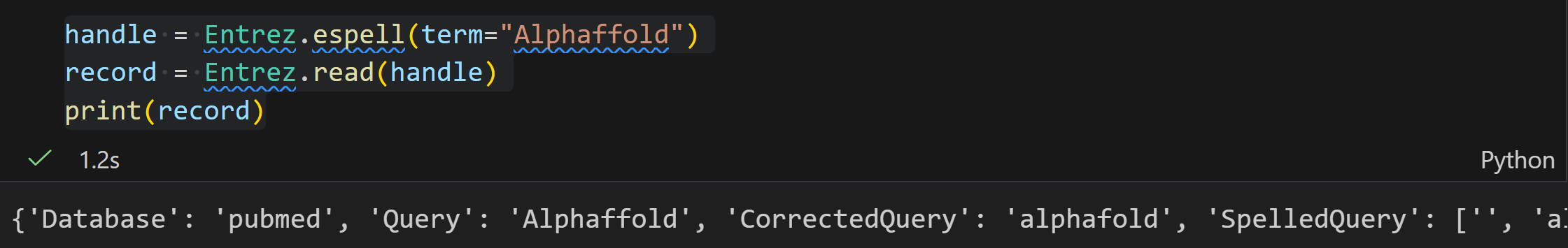
错误输入 vs 正确拼写
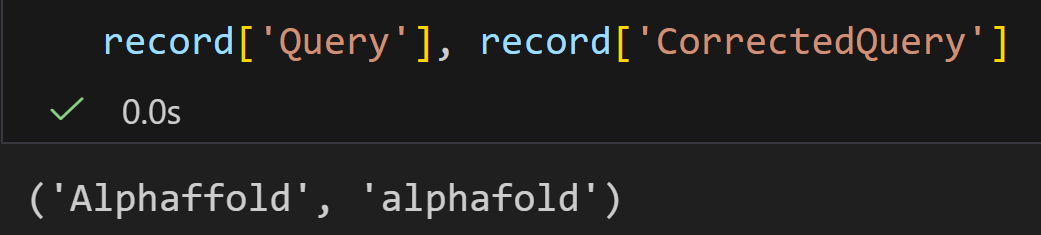
二,Entrez Direct
就像前面说的那样,BioPython中的Entrez模块是对Entrez Direct的套壳,
熟悉python的可以直接投入BioPython,或者用python自己的XML解析模块解析Entrez Direct返回的内容;熟悉Awk之类shell脚本语言的,可以直接上手Entrez Direct,其本身也有一套类似体系的脚本语言形式。
毕竟biopython是套壳,我们要获取第一手资料,还是得了解NCBI官方的实现工具。
因为详细的Entrez Programming Utilities一整套有很多要学,
和本篇博客相关的,我认为重要的只有Entrez Direct,核心看懂下面这篇即可(仅代表个人观点 ):
https://www.ncbi.nlm.nih.gov/books/NBK179288/

后续可能会出一篇讲这个EDirect脚本语言。