简述
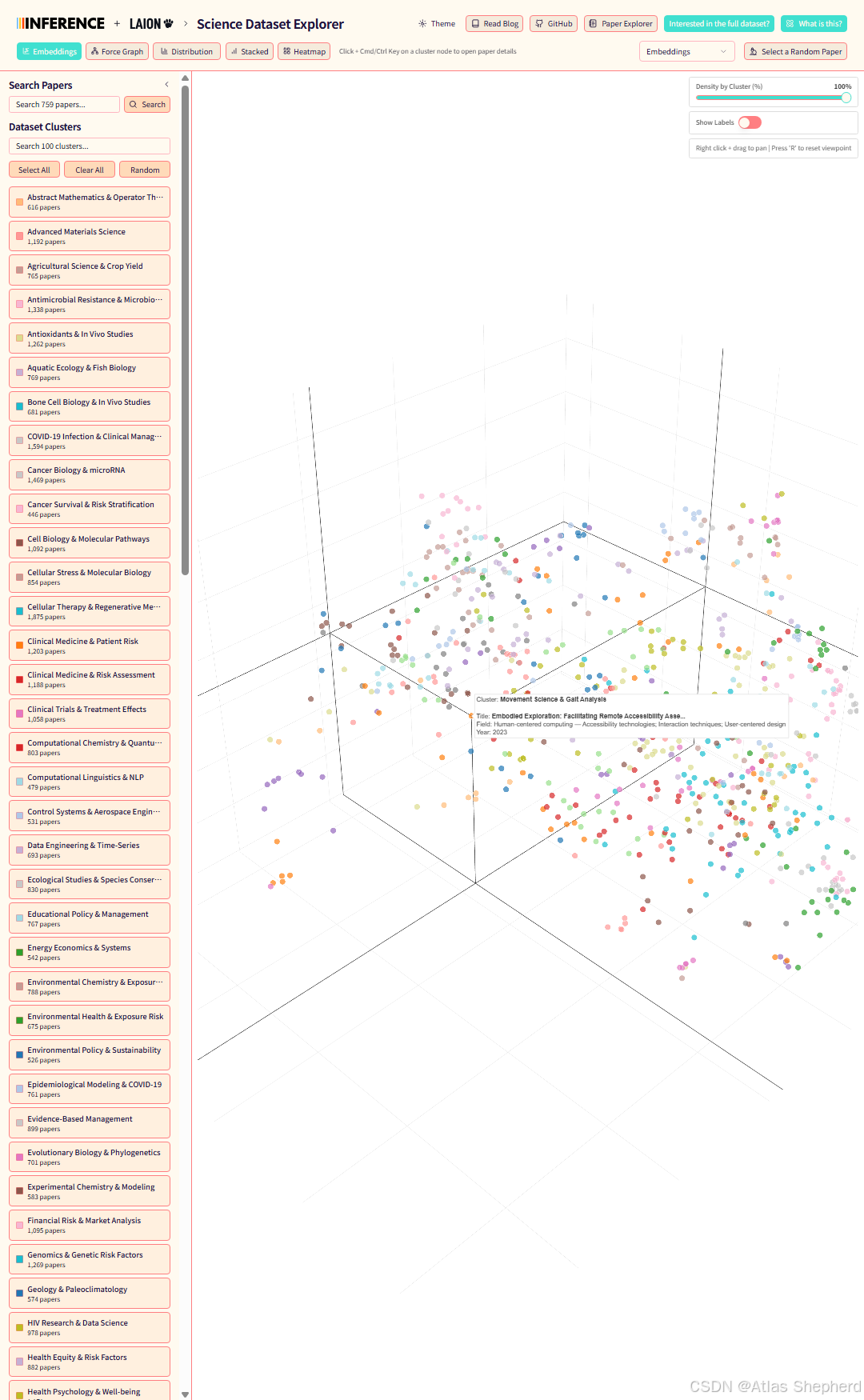
Aella Science Dataset Explorer 是一个科学论文数据集的交互式可视化探索工具,基于 React + FastAPI + SQLite 技术栈。项目由 Inference.net 和 LAION 合作开发。
环境要求
必需工具
-
**Python 3.11+** - 后端运行环境
-
uv - Python 包管理器
-
bun - JavaScript 运行时和包管理器
-
Task - 任务运行器
工具安装验证
# 验证安装
python --version # 应该显示 3.11.x 或更高
uv --version # 应该显示版本号
bun --version # 应该显示版本号
task --version # 应该显示版本号完整部署流程
1. 克隆项目
git clone https://gitcode.com/2301_76444133/aella.git
cd aella2. 安装工具(如未安装)
安装 uv
pip install uv安装 bun(Windows)
# 方法1:使用 PowerShell
powershell -c "irm bun.sh/install.ps1 | iex"
# 方法2:如果 PATH 未自动添加,手动添加
# 将 C:\Users\用户名\.bun\bin 添加到系统 PATH 环境变量安装 Task(Windows)
# 方法1:手动下载
# 访问 https://github.com/go-task/task/releases
# 下载 task_windows_amd64.zip,解压得到 task.exe
# 将 task.exe 放入系统 PATH 或项目根目录
# 方法2:使用 bun 安装
bun add -g @go-task/cli3. 安装项目依赖
task setup这个命令会:
-
安装后端 Python 依赖(通过 uv)
-
安装前端 JavaScript 依赖(通过 bun)
4. 下载数据库
task db:setup如果失败,手动下载:
mkdir backend\data
curl -L -o backend\data\db.sqlite "https://laion-data-assets.inference.net/db.sqlite"5. 启动后端服务器
task backend:dev如果失败,手动启动:
创建本地服务器文件 (backend/server.py):
# 在 backend 目录中创建完整服务器
import uvicorn
from fastapi import FastAPI, HTTPException, Query
from fastapi.middleware.cors import CORSMiddleware
import sqlite3
import json
import logging
from pathlib import Path
from typing import Optional, List, Dict, Any
from datetime import datetime
# 配置日志
logging.basicConfig(level=logging.INFO, format='%(asctime)s - %(levelname)s - %(message)s')
logger = logging.getLogger(__name__)
# 创建 FastAPI 应用
app = FastAPI(
title="Aella Science Dataset Explorer API",
description="API for exploring scientific papers with embeddings and clusters",
version="1.0.0"
)
# 启用 CORS
app.add_middleware(
CORSMiddleware,
allow_origins=["*"],
allow_credentials=True,
allow_methods=["*"],
allow_headers=["*"],
)
# 数据库路径
DB_PATH = Path("data/db.sqlite")
logger.info(f"Database: {DB_PATH.resolve()} (exists: {DB_PATH.exists()})")
# 预定义的聚类颜色
CLUSTER_COLORS = [
"#1f77b4", "#ff7f0e", "#2ca02c", "#d62728", "#9467bd",
"#8c564b", "#e377c2", "#7f7f7f", "#bcbd22", "#17becf",
"#aec7e8", "#ffbb78", "#98df8a", "#ff9896", "#c5b0d5",
"#c49c94", "#f7b6d2", "#c7c7c7", "#dbdb8d", "#9edae5"
]
def get_cluster_color(cluster_id: int) -> str:
"""获取聚类颜色"""
if cluster_id < 0:
return "#E8E8E8"
return CLUSTER_COLORS[cluster_id % len(CLUSTER_COLORS)]
@app.get("/")
async def root():
return {
"message": "Aella Science Dataset Explorer API",
"version": "1.0.0",
"docs": "/docs"
}
@app.get("/api/papers")
async def get_papers(
cluster_id: Optional[int] = Query(None, description="Filter by cluster ID"),
limit: Optional[int] = Query(1000, description="Limit number of results"),
sample_size: Optional[int] = Query(None, description="Sample N papers per cluster")
):
"""获取论文数据"""
try:
logger.info(f"Fetching papers: cluster_id={cluster_id}, limit={limit}, sample_size={sample_size}")
with sqlite3.connect(str(DB_PATH)) as conn:
conn.row_factory = sqlite3.Row
if sample_size is not None and cluster_id is None:
# 抽样每个聚类的论文
all_rows = conn.execute("""
SELECT id, title, x, y, z, cluster_id,
COALESCE(claude_label, cluster_label) as cluster_label,
field_subfield, publication_year, classification
FROM papers
WHERE x IS NOT NULL AND y IS NOT NULL AND cluster_id IS NOT NULL
ORDER BY cluster_id, publication_year DESC
LIMIT ?
""", (sample_size * 50,)).fetchall()
# 抽样逻辑
cluster_samples = {}
for row in all_rows:
cid = row["cluster_id"]
if cid not in cluster_samples:
cluster_samples[cid] = []
if len(cluster_samples[cid]) < sample_size:
cluster_samples[cid].append(dict(row))
rows = []
for cid in sorted(cluster_samples.keys()):
rows.extend(cluster_samples[cid])
else:
# 普通查询
query = """
SELECT id, title, x, y, z, cluster_id,
COALESCE(claude_label, cluster_label) as cluster_label,
field_subfield, publication_year, classification
FROM papers
WHERE x IS NOT NULL AND y IS NOT NULL
"""
params = []
if cluster_id is not None:
query += " AND cluster_id = ?"
params.append(cluster_id)
query += " ORDER BY id"
if limit is not None:
query += " LIMIT ?"
params.append(limit)
rows = [dict(row) for row in conn.execute(query, params).fetchall()]
logger.info(f"Returning {len(rows)} papers")
return {"papers": rows}
except Exception as e:
logger.error(f"Database error: {e}")
raise HTTPException(status_code=500, detail=str(e))
@app.get("/api/clusters")
async def get_clusters():
"""获取聚类信息"""
try:
logger.info("Fetching clusters")
with sqlite3.connect(str(DB_PATH)) as conn:
conn.row_factory = sqlite3.Row
rows = conn.execute("""
SELECT cluster_id,
COALESCE(claude_label, cluster_label) as cluster_label,
COUNT(*) as count
FROM papers
WHERE cluster_id IS NOT NULL
GROUP BY cluster_id, cluster_label
ORDER BY cluster_id
""").fetchall()
clusters = []
for row in rows:
cluster = dict(row)
cluster["color"] = get_cluster_color(row["cluster_id"])
clusters.append(cluster)
logger.info(f"Returning {len(clusters)} clusters")
return {"clusters": clusters}
except Exception as e:
logger.error(f"Database error: {e}")
raise HTTPException(status_code=500, detail=str(e))
@app.get("/api/temporal-data")
async def get_temporal_data(
min_year: int = Query(1990, description="Minimum publication year"),
max_year: int = Query(2025, description="Maximum publication year")
):
"""获取时间序列数据(按年份和聚类的论文数量)"""
try:
logger.info(f"Fetching temporal data: {min_year}-{max_year}")
with sqlite3.connect(str(DB_PATH)) as conn:
conn.row_factory = sqlite3.Row
rows = conn.execute("""
SELECT
cluster_id,
COALESCE(claude_label, cluster_label) as cluster_label,
publication_year,
COUNT(*) as count
FROM papers
WHERE cluster_id IS NOT NULL
AND publication_year IS NOT NULL
AND publication_year >= ?
AND publication_year <= ?
GROUP BY cluster_id, cluster_label, publication_year
ORDER BY cluster_id, publication_year
""", (min_year, max_year)).fetchall()
# 按聚类组织数据
cluster_data = {}
for row in rows:
cluster_id = row["cluster_id"]
if cluster_id not in cluster_data:
cluster_data[cluster_id] = {
"cluster_id": cluster_id,
"cluster_label": row["cluster_label"],
"color": get_cluster_color(cluster_id),
"temporal_data": []
}
cluster_data[cluster_id]["temporal_data"].append({
"year": row["publication_year"],
"count": row["count"]
})
# 转换为列表
result = list(cluster_data.values())
logger.info(f"Returning temporal data for {len(result)} clusters")
return {"clusters": result}
except Exception as e:
logger.error(f"Database error: {e}")
raise HTTPException(status_code=500, detail=str(e))
@app.get("/api/search")
async def search_papers(
q: str = Query(..., description="Search query"),
limit: int = Query(100, description="Maximum results")
):
"""搜索论文"""
try:
logger.info(f"Searching for: {q}")
with sqlite3.connect(str(DB_PATH)) as conn:
conn.row_factory = sqlite3.Row
rows = conn.execute("""
SELECT id, title, x, y, z, cluster_id,
COALESCE(claude_label, cluster_label) as cluster_label,
field_subfield, publication_year, classification
FROM papers
WHERE x IS NOT NULL AND y IS NOT NULL
AND (title LIKE ? OR field_subfield LIKE ?)
ORDER BY id
LIMIT ?
""", (f"%{q}%", f"%{q}%", limit)).fetchall()
logger.info(f"Found {len(rows)} papers for search: {q}")
return {"papers": [dict(row) for row in rows]}
except Exception as e:
logger.error(f"Search error: {e}")
raise HTTPException(status_code=500, detail=str(e))
@app.get("/api/stats")
async def get_stats():
"""获取统计数据"""
try:
logger.info("Fetching stats")
with sqlite3.connect(str(DB_PATH)) as conn:
cursor = conn.cursor()
cursor.execute("SELECT COUNT(*) FROM papers")
total = cursor.fetchone()[0]
cursor.execute("SELECT COUNT(*) FROM papers WHERE x IS NOT NULL AND y IS NOT NULL")
with_coords = cursor.fetchone()[0]
cursor.execute("SELECT COUNT(DISTINCT cluster_id) FROM papers WHERE cluster_id IS NOT NULL")
num_clusters = cursor.fetchone()[0]
return {
"total_papers": total,
"papers_with_coordinates": with_coords,
"num_clusters": num_clusters
}
except Exception as e:
logger.error(f"Database error: {e}")
raise HTTPException(status_code=500, detail=str(e))
@app.get("/health")
async def health():
return {"status": "ok"}
if __name__ == "__main__":
print("=" * 60)
print("Aella Science Dataset Explorer - Complete Backend Server")
print("=" * 60)
print(f"Database: {DB_PATH.resolve()}")
print(f"API URL: http://localhost:8787")
print(f"API Docs: http://localhost:8787/docs")
print(f"Frontend: http://localhost:5173")
print("=" * 60)
uvicorn.run(
app,
host="0.0.0.0",
port=8787,
reload=False,
log_level="info"
)
EOF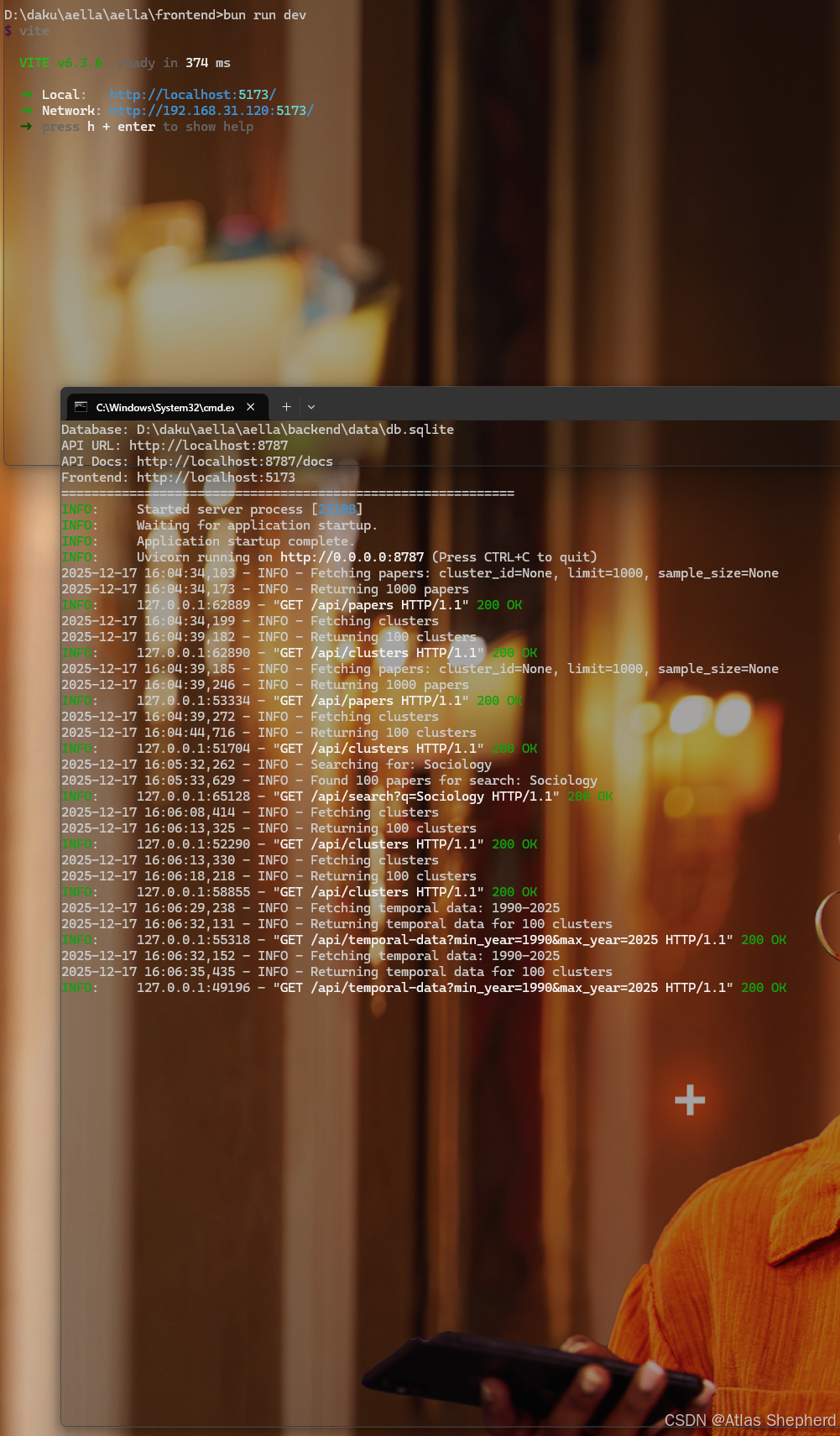
启动服务器:
cd backend
uv run python server.py6. 启动前端服务器
打开新的命令行窗口:
cd frontend
bun run dev7. 访问应用
-
后端 API:http://localhost:8787
-
API 文档:http://localhost:8787/docs
关键问题与解决方案
问题1:bun 安装后命令未找到
症状 :bun --version返回 "'bun' 不是内部或外部命令"
解决方案:
-
检查安装路径:
C:\Users\用户名\.bun\bin\bun.exe -
手动添加到 PATH 环境变量
-
重新启动命令行窗口
问题2:Task 命令未找到
症状 :task --version返回命令未找到
解决方案:
-
将 task.exe 放入系统 PATH 目录
-
或在项目根目录使用
.\task.exe(如果 task.exe 在项目目录中)
问题3:后端 Cloudflare Workers 依赖错误
症状 :ModuleNotFoundError: No module named 'workers'
解决方案:
-
创建本地 FastAPI 服务器,避免 Cloudflare Workers 特定依赖
-
使用标准的 SQLite 数据库连接
问题4:前端代理错误
症状:前端控制台显示代理连接错误
解决方案:
-
确保后端服务器在 8787 端口运行
-
检查后端 API 接口是否完整实现
-
修改前端代理配置(如有必要)
问题5:数据库下载失败
症状 :task db:setup执行失败
解决方案:
-
手动创建目录:
mkdir backend\data -
手动下载数据库文件
-
检查网络连接和防火墙设置
部署验证清单
后端验证
-
\] 服务器启动成功(端口 8787)
-
\] `/api/papers`返回论文数据
-
\] 数据库连接正常
-
\] 开发服务器启动成功(端口 5173)
-
\] 能够显示论文数据点
完整功能验证
-
\] 论文数据可视化显示
-
\] 交互功能正常(缩放、拖拽)
故障排除指南
快速诊断命令
# 检查后端状态
curl http://localhost:8787/api/papers?limit=1
# 检查前端状态
curl http://localhost:5173
# 检查数据库
python -c "import sqlite3; conn=sqlite3.connect('backend/data/db.sqlite'); print('Tables:', conn.execute('SELECT name FROM sqlite_master WHERE type=\"table\"').fetchall())"常见错误日志分析
-
端口占用:修改端口号或关闭占用程序
-
文件权限:以管理员身份运行命令行
-
依赖冲突:重新安装依赖或使用虚拟环境
-
网络问题:检查代理设置和防火墙
生产环境部署建议
后端优化
-
使用 Gunicorn 或 Uvicorn 工作进程
-
配置反向代理(Nginx)
-
启用 Gzip 压缩
-
设置合适的 CORS 策略
前端优化
-
构建生产版本:
bun run build -
配置静态文件服务
-
启用缓存策略
-
设置 CDN 加速
项目结构说明
aella/
├── backend/ # FastAPI 后端
│ ├── data/ # 数据库文件
│ ├── src/ # 源代码
│ └── server.py # 本地服务器入口
├── frontend/ # React 前端
│ ├── src/ # 前端源代码
│ └── package.json # 前端依赖
└── Taskfile.yml # 任务配置总结
Aella Science Dataset Explorer 的部署过程相对直接,主要挑战在于环境配置和工具安装。通过本笔记的步骤,可以系统性地完成从零到生产的部署。关键成功因素包括正确的工具版本、完整的依赖安装和适当的本地服务器配置。
部署完成后,用户可以通过直观的可视化界面探索科学论文数据集,支持按聚类筛选、时间序列分析等功能,为科研数据探索提供了强大的工具支持。