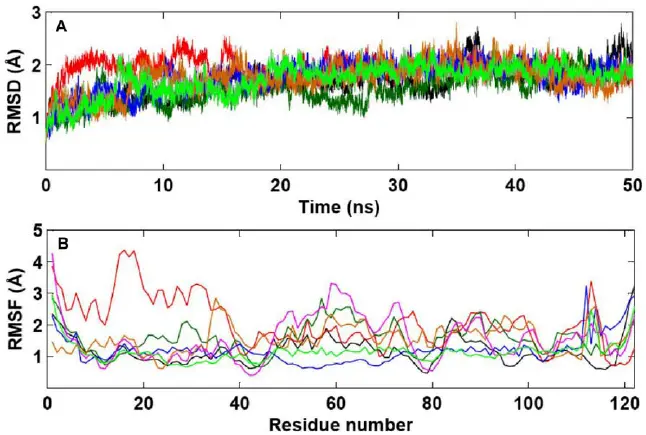
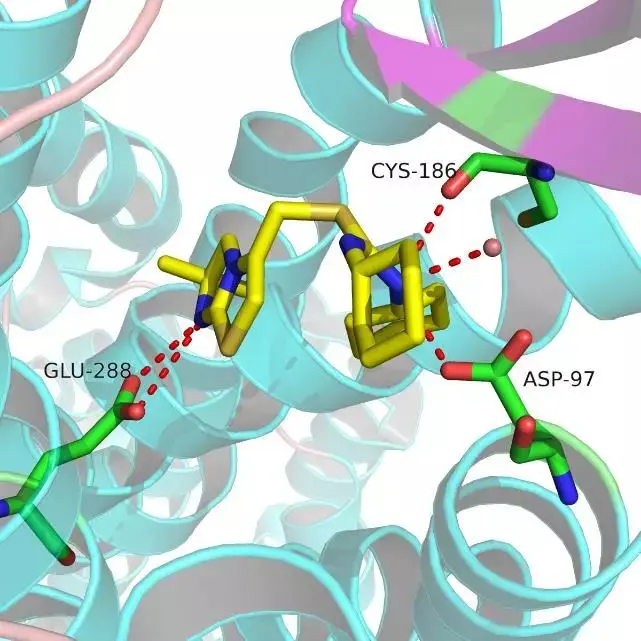
复制代码
#!/usr/bin/python
####zhaoyunfei
####20260113
#!/usr/bin/env python3
"""
PyRosetta分子对接 - 直接生成PyMOL可视化文件
生成可直接导入PyMOL的构象文件和RMSD计算结果
"""
import os
import sys
import numpy as np
import pandas as pd
import warnings
warnings.filterwarnings('ignore')
from datetime import datetime
import subprocess
import shutil
# ==================== 配置部分 ====================
# 输入文件
PROTEIN_PDB_FILE = "1ake.pdb" # 蛋白PDB文件
LIGAND_FILE = "ligand.sdf" # 配体文件
LIGAND_FORMAT = "sdf" # 配体文件格式
# 工作目录
TIMESTAMP = datetime.now().strftime("%Y%m%d_%H%M%S")
WORK_DIR = f"./docking_pymol_{TIMESTAMP}"
# 对接参数
DOCKING_CHAIN = "A" # 蛋白链ID
LIGAND_RESNAME = "LIG" # 配体残基名
NUM_POSES = 10 # 生成10个构象
# ==================== 初始化 PyRosetta ====================
print("=" * 80)
print("PyRosetta分子对接 - 生成PyMOL可视化文件")
print(f"开始时间: {datetime.now().strftime('%Y-%m-%d %H:%M:%S')}")
print("=" * 80)
try:
import pyrosetta
from pyrosetta import Pose, pose_from_pdb
from pyrosetta.rosetta.core.pose import append_pose_to_pose
from pyrosetta.rosetta.core.import_pose import pose_from_file
from pyrosetta.rosetta.protocols.docking import setup_foldtree, DockMCMProtocol
from pyrosetta.rosetta.core.scoring import ScoreFunctionFactory
from pyrosetta.rosetta.core.scoring import all_atom_rmsd
# 初始化PyRosetta
pyrosetta.init(extra_options="-mute all -ignore_unrecognized_res true -ex1 -ex2aro")
print("✓ PyRosetta 初始化成功")
except ImportError as e:
print(f"✗ PyRosetta 导入错误: {e}")
sys.exit(1)
# ==================== 检查输入文件 ====================
print("\n1. 检查输入文件...")
if not os.path.exists(PROTEIN_PDB_FILE):
print(f"✗ 错误:蛋白文件不存在: {PROTEIN_PDB_FILE}")
sys.exit(1)
print(f" ✓ 蛋白文件: {PROTEIN_PDB_FILE}")
if not os.path.exists(LIGAND_FILE):
print(f"✗ 错误:配体文件不存在: {LIGAND_FILE}")
sys.exit(1)
print(f" ✓ 配体文件: {LIGAND_FILE}")
# ==================== 创建输出目录 ====================
print("\n2. 创建工作目录...")
PYMOL_DIR = os.path.join(WORK_DIR, "pymol_files")
STRUCT_DIR = os.path.join(WORK_DIR, "structures")
os.makedirs(WORK_DIR, exist_ok=True)
os.makedirs(PYMOL_DIR, exist_ok=True)
os.makedirs(STRUCT_DIR, exist_ok=True)
print(f" 工作目录: {WORK_DIR}")
print(f" PyMOL文件目录: {PYMOL_DIR}")
# ==================== 配体文件格式转换 ====================
print(f"\n3. 转换配体文件格式...")
ligand_pdb_file = os.path.join(WORK_DIR, "ligand.pdb")
if LIGAND_FORMAT.lower() != 'pdb':
try:
# 尝试使用Open Babel
cmd = ['obabel', LIGAND_FILE, '-i', LIGAND_FORMAT.lower(), '-o', 'pdb', '-O', ligand_pdb_file, '-h']
result = subprocess.run(cmd, capture_output=True, text=True, timeout=30)
if result.returncode == 0:
print(f" ✓ Open Babel转换成功")
else:
# 尝试使用RDKit
from rdkit import Chem
from rdkit.Chem import AllChem
if LIGAND_FORMAT.lower() == 'sdf':
mol = Chem.MolFromMolFile(LIGAND_FILE)
elif LIGAND_FORMAT.lower() == 'mol2':
mol = Chem.MolFromMol2File(LIGAND_FILE)
elif LIGAND_FORMAT.lower() == 'mol':
mol = Chem.MolFromMolFile(LIGAND_FILE)
elif LIGAND_FORMAT.lower() == 'pdb':
mol = Chem.MolFromPDBFile(LIGAND_FILE)
else:
print(f" ✗ 不支持的格式")
sys.exit(1)
if mol:
mol = Chem.AddHs(mol)
AllChem.EmbedMolecule(mol, randomSeed=42)
AllChem.MMFFOptimizeMolecule(mol)
Chem.MolToPDBFile(mol, ligand_pdb_file)
print(f" ✓ RDKit转换成功")
except:
print(f" ✗ 格式转换失败")
sys.exit(1)
else:
shutil.copy(LIGAND_FILE, ligand_pdb_file)
# ==================== 加载蛋白质和配体 ====================
print(f"\n4. 加载蛋白质和配体...")
try:
# 加载蛋白质
protein_pose = pose_from_pdb(PROTEIN_PDB_FILE)
print(f" ✓ 蛋白质: {protein_pose.total_residue()}个残基")
# 加载配体
ligand_pose = pose_from_file(ligand_pdb_file)
print(f" ✓ 配体: {ligand_pose.total_residue()}个残基")
except Exception as e:
print(f"✗ 加载失败: {e}")
sys.exit(1)
# ==================== 执行对接 ====================
print(f"\n5. 执行分子对接...")
print(f" 生成{NUM_POSES}个构象...")
# 创建复合物
complex_pose = protein_pose.clone()
append_pose_to_pose(complex_pose, ligand_pose, new_chain=True)
ligand_chain_id = complex_pose.num_chains()
# 设置对接
scorefxn = ScoreFunctionFactory.create_score_function('ref2015')
docking_scorefxn = ScoreFunctionFactory.create_score_function('docking')
setup_foldtree(complex_pose, f"{ligand_chain_id}", pyrosetta.rosetta.utility.vector1_int([1]))
dock_mcm = DockMCMProtocol(ligand_chain_id)
dock_mcm.set_scorefxn(docking_scorefxn)
dock_mcm.set_partners(f"{ligand_chain_id}")
dock_mcm.set_outer_cycles(10)
# 存储对接结果
all_poses = []
energies = []
for i in range(NUM_POSES):
print(f" 对接 {i+1}/{NUM_POSES}...", end="\r")
dock_pose = complex_pose.clone()
dock_mcm.apply(dock_pose)
# 计算能量
total_score = scorefxn(dock_pose)
# 保存PDB文件
pdb_file = os.path.join(STRUCT_DIR, f"pose_{i+1:03d}.pdb")
dock_pose.dump_pdb(pdb_file)
all_poses.append(dock_pose)
energies.append(total_score)
print(f"\n ✓ 对接完成")
# ==================== 选择最佳构象 ====================
print(f"\n6. 选择最佳构象...")
best_idx = np.argmin(energies)
best_pose_id = best_idx + 1
best_energy = energies[best_idx]
best_pose = all_poses[best_idx]
print(f" ✓ 最佳构象: Pose {best_pose_id}")
print(f" ✓ 最佳能量: {best_energy:.2f} REU")
# ==================== 计算RMSD ====================
print(f"\n7. 计算RMSD...")
# 获取配体残基索引
ligand_start = protein_pose.total_residue() + 1
ligand_end = best_pose.total_residue()
ligand_residues = list(range(ligand_start, ligand_end + 1))
# 计算每个构象相对于最佳构象的RMSD
rmsd_values = []
for i, pose in enumerate(all_poses):
if i == best_idx:
rmsd = 0.0
else:
rmsd = all_atom_rmsd(best_pose, pose, ligand_residues, ligand_residues)
rmsd_values.append(rmsd)
# 创建结果表格
results_df = pd.DataFrame({
'Pose_ID': list(range(1, NUM_POSES + 1)),
'Energy_REU': energies,
'RMSD_A': rmsd_values
})
# 保存RMSD结果
rmsd_csv = os.path.join(PYMOL_DIR, "rmsd_results.csv")
results_df.to_csv(rmsd_csv, index=False)
print(f" ✓ RMSD计算完成")
print(f" ✓ 保存RMSD结果: {rmsd_csv}")
print(f"\n RMSD结果:")
print(results_df.to_string(index=False))
# ==================== 生成PyMOL可视化文件 ====================
print(f"\n8. 生成PyMOL可视化文件...")
# 1. 生成复合PDB文件(包含所有构象)
print(" 生成复合PDB文件...")
composite_pdb = os.path.join(PYMOL_DIR, "all_poses_composite.pdb")
with open(composite_pdb, 'w') as out_f:
for i in range(NUM_POSES):
pose_id = i + 1
pose_file = os.path.join(STRUCT_DIR, f"pose_{pose_id:03d}.pdb")
with open(pose_file, 'r') as in_f:
lines = in_f.readlines()
# 写入MODEL记录
out_f.write(f"MODEL {pose_id}\n")
out_f.write(f"REMARK Pose {pose_id}\n")
out_f.write(f"REMARK Energy: {energies[i]:.2f} REU\n")
out_f.write(f"REMARK RMSD to best: {rmsd_values[i]:.3f} Å\n")
# 写入原子记录
for line in lines:
if line.startswith('ATOM') or line.startswith('HETATM'):
out_f.write(line)
out_f.write("ENDMDL\n")
print(f" ✓ 复合PDB文件: {composite_pdb}")
# 2. 生成最佳构象单独文件
print(" 生成最佳构象文件...")
best_pdb = os.path.join(PYMOL_DIR, "best_pose.pdb")
shutil.copy(os.path.join(STRUCT_DIR, f"pose_{best_pose_id:03d}.pdb"), best_pdb)
print(f" ✓ 最佳构象文件: {best_pdb}")
# 3. 生成PyMOL会话文件
print(" 生成PyMOL会话文件...")
pymol_script = os.path.join(PYMOL_DIR, "view_docking.pml")
script_content = f"""# PyMOL对接可视化脚本
# 生成时间: {datetime.now().strftime('%Y-%m-%d %H:%M:%S')}
# 清除场景
cmd.delete("all")
# 加载复合PDB文件(包含所有构象)
cmd.load("{os.path.basename(composite_pdb)}", "all_poses")
# 设置显示
cmd.hide("everything", "all_poses")
cmd.show("sticks", "all_poses and resn {LIGAND_RESNAME}")
# 按能量着色(红->蓝渐变)
cmd.util.cbay("all_poses and resn {LIGAND_RESNAME}")
# 加载蛋白质(显示一次)
protein_file = "{os.path.basename(os.path.join(WORK_DIR, 'protein.pdb'))}"
cmd.load(protein_file, "protein")
cmd.hide("everything", "protein")
cmd.show("cartoon", "protein")
cmd.color("gray", "protein")
cmd.set("cartoon_transparency", 0.5)
# 对齐所有构象到最佳构象
cmd.select("ref_ligand", f"all_poses and resn {LIGAND_RESNAME} and model {best_pose_id}")
for i in range(1, {NUM_POSES}+1):
if i != {best_pose_id}:
cmd.select(f"lig_{{i}}", f"all_poses and resn {LIGAND_RESNAME} and model {{i}}")
cmd.align(f"lig_{{i}}", "ref_ligand")
# 设置视角
cmd.zoom("protein", 10)
# 显示标签
cmd.set("label_color", "white")
cmd.set("label_size", 14)
# 为每个构象添加标签
for i in range(1, {NUM_POSES}+1):
cmd.label(f"all_poses and model {{i}} and name C1",
f"Pose {{i}}\\nE: {energies[i-1]:.1f}\\nRMSD: {rmsd_values[i-1]:.2f}Å")
print("="*60)
print("对接构象可视化加载完成!")
print("="*60)
print("显示内容:")
print("1. 蛋白质: 灰色卡通图")
print("2. 配体构象: 彩虹色棍棒图")
print("3. 标签显示: 构象编号、能量、RMSD")
print()
print("操作提示:")
print("• 按空格键切换构象")
print("• 使用'movie'命令创建动画")
print("• 输入'show surface, protein'显示蛋白质表面")
print("="*60)
"""
with open(pymol_script, 'w') as f:
f.write(script_content)
print(f" ✓ PyMOL脚本: {pymol_script}")
# 4. 生成蛋白质PDB文件
protein_pdb = os.path.join(PYMOL_DIR, "protein.pdb")
protein_pose.dump_pdb(protein_pdb)
print(f" ✓ 蛋白质文件: {protein_pdb}")
# 5. 生成能量-RMSD数据文件(用于PyMOL)
print(" 生成PyMOL数据文件...")
pymol_data = os.path.join(PYMOL_DIR, "docking_data.txt")
with open(pymol_data, 'w') as f:
f.write("# PyMOL对接数据文件\n")
f.write(f"# 生成时间: {datetime.now().strftime('%Y-%m-%d %H:%M:%S')}\n")
f.write("# 列: Pose_ID, Energy_REU, RMSD_A\n")
f.write("\n")
for i in range(NUM_POSES):
f.write(f"{i+1} {energies[i]:.2f} {rmsd_values[i]:.3f}\n")
print(f" ✓ 数据文件: {pymol_data}")
# 6. 生成一键加载脚本
print(" 生成一键加载脚本...")
load_script = os.path.join(PYMOL_DIR, "load_all.py")
load_content = f"""#!/usr/bin/env python
# PyMOL一键加载脚本
import os
print("正在加载对接结果...")
# 加载所有文件
cmd.load("{os.path.basename(composite_pdb)}", "docking_poses")
cmd.load("{os.path.basename(protein_pdb)}", "protein")
# 设置显示
cmd.hide("everything")
cmd.show("cartoon", "protein")
cmd.show("sticks", "docking_poses and resn {LIGAND_RESNAME}")
# 着色
cmd.color("gray", "protein")
cmd.util.chainbow("docking_poses and resn {LIGAND_RESNAME}")
# 设置透明度
cmd.set("cartoon_transparency", 0.3, "protein")
cmd.set("stick_transparency", 0.2, "docking_poses")
print("加载完成!")
print(f"共加载{{NUM_POSES}}个对接构象")
print(f"最佳构象: Pose {{best_pose_id}} (能量: {{best_energy:.2f}} REU)")
"""
with open(load_script, 'w') as f:
f.write(load_content)
# 7. 生成README文件
readme_file = os.path.join(PYMOL_DIR, "README.txt")
with open(readme_file, 'w') as f:
f.write(f"""PyMOL对接可视化文件使用说明
============================================
生成时间: {datetime.now().strftime('%Y-%m-%d %H:%M:%S')}
工作目录: {WORK_DIR}
文件清单:
1. all_poses_composite.pdb - 包含所有对接构象的复合PDB文件
2. best_pose.pdb - 最佳对接构象单独文件
3. protein.pdb - 蛋白质结构文件
4. view_docking.pml - PyMOL可视化脚本
5. load_all.py - 一键加载脚本
6. rmsd_results.csv - RMSD计算结果
7. docking_data.txt - 对接数据文件
使用方法:
方法1: 使用PyMOL脚本
pymol view_docking.pml
方法2: 手动加载
pymol all_poses_composite.pdb protein.pdb
然后在PyMOL命令行输入:
run load_all.py
方法3: 只查看最佳构象
pymol best_pose.pdb protein.pdb
可视化说明:
- 蛋白质显示为灰色卡通图
- 对接构象显示为彩虹色棍棒图(按能量着色)
- 红色表示低能量(好),蓝色表示高能量(差)
- 按空格键可以切换不同构象
对接结果概览:
- 总构象数: {NUM_POSES}
- 最佳构象: Pose {best_pose_id}
- 最佳能量: {best_energy:.2f} REU
- 平均RMSD: {np.mean(rmsd_values):.3f} Å
- RMSD范围: {np.min(rmsd_values):.3f} - {np.max(rmsd_values):.3f} Å
PyMOL常用命令:
show surface, protein # 显示蛋白质表面
distance hbonds, ligand, protein, 3.5 # 显示氢键
select pocket, protein within 5 of resn {LIGAND_RESNAME} # 选择结合口袋
show sticks, pocket # 显示口袋残基
movie # 创建构象动画
""")
print(f" ✓ README文件: {readme_file}")
# 8. 生成RMSD-能量散点图(PNG格式,也可在PyMOL中查看)
print(" 生成RMSD-能量散点图...")
import matplotlib.pyplot as plt
plt.figure(figsize=(8, 6))
plt.scatter(rmsd_values, energies, c=range(NUM_POSES), cmap='viridis', s=100, alpha=0.8, edgecolors='black')
# 标记最佳构象
plt.scatter(rmsd_values[best_idx], energies[best_idx],
color='red', s=200, marker='*', label=f'Best Pose {best_pose_id}')
plt.xlabel('RMSD to Best Pose (Å)', fontsize=12)
plt.ylabel('Energy (REU)', fontsize=12)
plt.title(f'Docking Results: Energy vs RMSD (N={NUM_POSES})', fontsize=14)
plt.colorbar(label='Pose ID')
plt.grid(True, alpha=0.3)
plt.legend()
# 添加统计信息
stats_text = f'Best: Pose {best_pose_id}\\nEnergy: {best_energy:.2f} REU\\nAvg RMSD: {np.mean(rmsd_values):.2f} Å'
plt.text(0.02, 0.98, stats_text, transform=plt.gca().transAxes,
fontsize=10, verticalalignment='top',
bbox=dict(boxstyle='round', facecolor='wheat', alpha=0.5))
plot_file = os.path.join(PYMOL_DIR, "energy_vs_rmsd.png")
plt.tight_layout()
plt.savefig(plot_file, dpi=300)
plt.close()
print(f" ✓ 散点图: {plot_file}")
# ==================== 复制所有文件到PyMOL目录 ====================
print(f"\n9. 整理文件...")
# 复制所有结构文件
for i in range(NUM_POSES):
src = os.path.join(STRUCT_DIR, f"pose_{i+1:03d}.pdb")
dst = os.path.join(PYMOL_DIR, f"pose_{i+1:03d}_single.pdb")
shutil.copy(src, dst)
# 复制原始蛋白文件
shutil.copy(PROTEIN_PDB_FILE, os.path.join(PYMOL_DIR, "original_protein.pdb"))
print(f" ✓ 所有文件已整理到: {PYMOL_DIR}")
# ==================== 生成使用说明 ====================
print(f"\n10. 生成使用说明...")
final_readme = os.path.join(WORK_DIR, "USAGE.txt")
with open(final_readme, 'w') as f:
f.write(f"""PyRosetta对接结果使用说明
============================================
分析完成时间: {datetime.now().strftime('%Y-%m-%d %H:%M:%S')}
重要文件位置:
1. PyMOL可视化文件目录: {PYMOL_DIR}/
2. 原始对接结构: {STRUCT_DIR}/
3. 分析结果: {WORK_DIR}/
快速开始:
1. 打开PyMOL
2. 执行以下任一命令:
# 方法1: 使用脚本(推荐)
run {PYMOL_DIR}/view_docking.pml
# 方法2: 加载复合文件
load {PYMOL_DIR}/all_poses_composite.pdb
load {PYMOL_DIR}/protein.pdb
run {PYMOL_DIR}/load_all.py
# 方法3: 只查看最佳构象
load {PYMOL_DIR}/best_pose.pdb
load {PYMOL_DIR}/protein.pdb
对接结果摘要:
- 对接构象总数: {NUM_POSES}
- 最佳构象编号: Pose {best_pose_id}
- 最佳构象能量: {best_energy:.2f} REU
- 相对于最佳构象的平均RMSD: {np.mean(rmsd_values):.3f} Å
- 详细的RMSD结果: {PYMOL_DIR}/rmsd_results.csv
PyMOL操作提示:
1. 查看不同构象: 按空格键
2. 显示蛋白质表面: show surface, protein
3. 显示氢键相互作用: distance hbonds, resn LIG, protein, 3.5
4. 创建构象动画: movie
5. 保存当前视图: File -> Save Session
文件说明:
{'-'*50}
all_poses_composite.pdb - 所有构象的复合文件(推荐)
best_pose.pdb - 最佳构象单独文件
protein.pdb - 处理后的蛋白质结构
view_docking.pml - 完整可视化脚本
load_all.py - 一键加载脚本
energy_vs_rmsd.png - RMSD-能量关系图
rmsd_results.csv - 详细的RMSD数据
pose_XXX_single.pdb - 单个构象文件
============================================
所有文件已准备就绪,可直接在PyMOL中使用!
""")
print(f" ✓ 使用说明: {final_readme}")
# ==================== 完成 ====================
print("\n" + "=" * 80)
print("PyMOL可视化文件生成完成!")
print("=" * 80)
print(f"\n生成的文件:")
print(f" ✓ {PYMOL_DIR}/all_poses_composite.pdb (主要文件)")
print(f" ✓ {PYMOL_DIR}/best_pose.pdb (最佳构象)")
print(f" ✓ {PYMOL_DIR}/protein.pdb (蛋白质)")
print(f" ✓ {PYMOL_DIR}/view_docking.pml (PyMOL脚本)")
print(f" ✓ {PYMOL_DIR}/energy_vs_rmsd.png (关系图)")
print(f" ✓ {PYMOL_DIR}/rmsd_results.csv (RMSD数据)")
print(f"\n使用方法:")
print(f" 1. 打开PyMOL")
print(f" 2. 执行: run {PYMOL_DIR}/view_docking.pml")
print(f" 或: load {PYMOL_DIR}/all_poses_composite.pdb")
print(f" load {PYMOL_DIR}/protein.pdb")
print(f"\n对接结果:")
print(f" • 最佳构象: Pose {best_pose_id}")
print(f" • 最佳能量: {best_energy:.2f} REU")
print(f" • 平均RMSD: {np.mean(rmsd_values):.3f} Å")
print(f"\n详细说明:")
print(f" ✓ {WORK_DIR}/USAGE.txt")
print(f"\n结束时间: {datetime.now().strftime('%Y-%m-%d %H:%M:%S')}")
print("=" * 80)