数据和代码获取:请查看主页个人信息!!!
大家好!欢迎来到R语言数据分析视界。相信大家对火山图的绘制方法已经并不陌生,我们前面的推文也有介绍过。
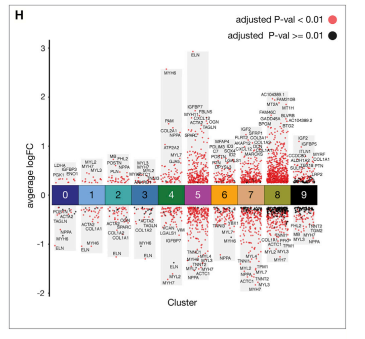
火山图适合展示两个分组之间的差异基因/代谢物等信息;然而,我们在进行实验设计时,往往会纳入多个分组。如果我们想看所有分组之间差异情况,就需要绘制多个火山图。今天我将向大家介绍如何使用ggplot2绘制多组火山图,用于同时展示多组之间差异情况。

相关性热图样式灵感来源于Cell杂志的一篇文章;绘图代码参考:https://zhuanlan.zhihu.com/p/516955474

接下来我们来进行分析和可视化展示,首先载入本次绘图数据:
Step1:数据载入
rm(list=ls())pacman::p_load(tidyverse, ggrepel, reshape2, rio)# 载入数据DEG_limma_voom <- read.table("DEG_limma_voom.txt", header=T, sep="\t")接下来,我们对数据进行处理,剔除不显著的数据。我们使用mutate函数创建一个新的列"change",根据给定的条件给基因分类为"Up"(上调)或"Down"(下调),并过滤掉"No change"的基因。
Step2:定义分组,剔除不显著的基因
# 剔除不显著的数据d1 <- DEG_limma_voom %>% mutate(change = as.factor(ifelse(P.Value < 0.05 & abs(logFC) > 1, ifelse(logFC > 1 ,'Up','Down'),'No change'))) %>% filter(change != 'No change') %>% select(-change)
# 这里我们创建一个火山图分组set.seed(123)g <- rep(paste0('Volcano_', 1:10),length.out = nrow(d1)) %>% .[sample(nrow(d1))]g[1:50]
# 定义P值分组data <- d1 %>% rownames_to_column('gene') %>% mutate(group = g) %>% mutate(label = ifelse(adj.P.Val < 0.001,"adjust P-val<0.001","adjust P-val >= 0.001"))
table(data$group)table(data$label)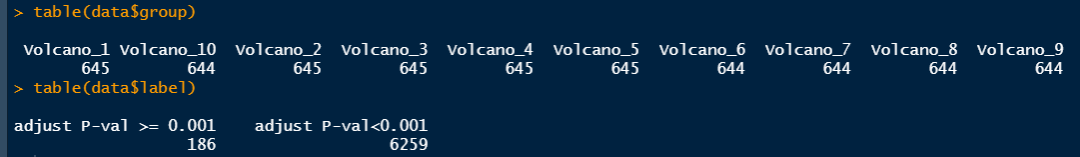
为了突出显示每个火山图分组中最显著的差异基因,我们使用top_n函数,选取每个分组中表达差异最显著的10个基因。
Step3:获取每个火山图中前十个差异基因
#获取每个group中表达差异最显著的10个基因,根据logFC的绝对值;TopGene <- data %>% group_by(group) %>% distinct(gene, .keep_all = T) %>% top_n(10, abs(logFC))table(TopGene$group)TopGene$gene
接下来,我们准备背景柱状图的数据。这些数据用于绘制背景柱状图,以突出显示每个分组的表达差异范围。
Step4:背景柱状图数据准备
# 背景柱状图数据准备dbar <- data %>% group_by(group) %>% summarise_all(list(min = min, max = max)) %>% select(group, logFC_min, logFC_max, label_min) %>% rename(label = label_min)dbar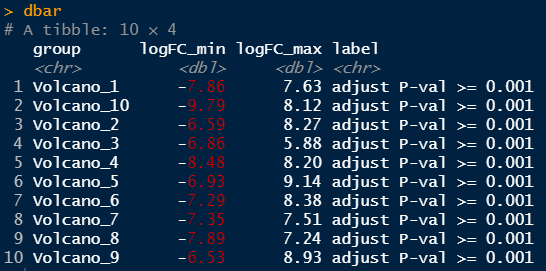
Step5:绘图
# 直接出图ggplot()+ geom_col(data = dbar, # 绘制负向背景柱状图 mapping = aes(x = group,y = logFC_min), fill = "#dcdcdc",alpha = 0.6, width = 0.7) + geom_col(data = dbar, # 绘制正向背景柱状图 mapping = aes(x = group,y = logFC_max), fill = "#dcdcdc",alpha = 0.6, width = 0.7) + geom_jitter(data = data, # 绘制所有数据点 aes(x = group, y = logFC, color = label), size = 0.85, width =0.3) + geom_jitter(data = TopGene, # 绘制top10数据点 aes(x = group, y = logFC, color = label), size = 1, width =0.35) + geom_tile(data = TopGene, # 绘制中心分组标记图 aes(x = group, y = 0, fill = group), height=1.5, color = "black", alpha = 0.6, show.legend = F) + ggsci::scale_fill_npg() + # 自定义颜色 ggsci::scale_color_npg() + # 自定义颜色 geom_text_repel(data = filter(data, gene %in% TopGene$gene), # 这里的filter很关键,筛选你想要标记的基因 aes(x = group, y = logFC, label = gene), size = 2, max.overlaps = getOption("ggrepel.max.overlaps", default = 15), color = 'black', force = 1.2, arrow = arrow(length = unit(0.008, "npc"), type = "open", ends = "last")) + labs(x="Cluster", y="Average logFC") + geom_text(data=TopGene, # 绘制中心分组标记图文本注释 aes(x=group, y=0, label=group), size = 3, color ="white") + theme_minimal() + theme(axis.title = element_text(size = 13,color = "black",face = "bold"), axis.line.y = element_line(color = "black",size = 1.2), axis.line.x = element_blank(), axis.text.x = element_blank(), panel.grid = element_blank(), legend.position = "top", legend.direction = "vertical", legend.justification = c(1,0), legend.text = element_text(size = 13))ggsave('pic.png', width = 10, height = 6, bg = 'white')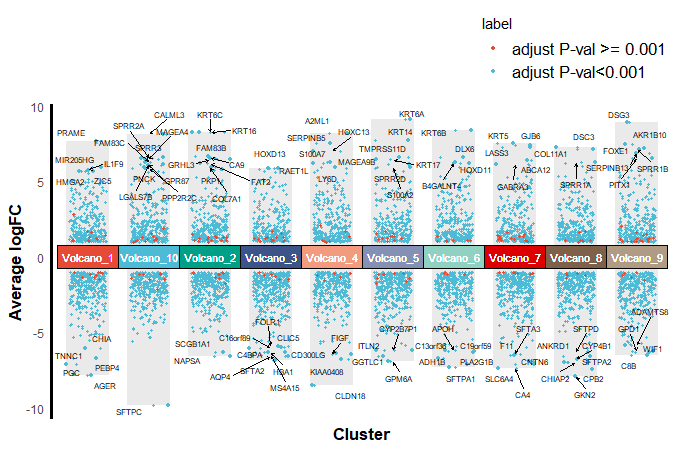
关键词"多组火山图" 获得本期代码和数据。