遗传算法与深度学习实战(37)------NEAT初体验
0. 前言
NEAT-Python封装了许多优化模式的工具,包括网络超参数、架构和参数优化以及增加拓扑结构。在本节中,我们将使用 sklearn 库构建示例数据集可视化 NEAT 分类结果。
1. 数据集加载
生成分类问题数据集,生成一个相对容易分类的数据集:
python
import numpy as np
import sklearn
import sklearn.datasets
import sklearn.linear_model
import matplotlib.pyplot as plt
from IPython.display import clear_output
import time
import random
import neat
number_samples = 10 #@param {type:"slider", min:10, max:1000, step:5}
difficulty = 1 #@param {type:"slider", min:1, max:5, step:1}
problem = "circles" #@param ["classification", "blobs", "gaussian quantiles", "moons", "circles"]
number_features = 2
number_classes = 2
middle_layer = 5 #@param {type:"slider", min:5, max:25, step:1}
def load_data(problem):
if problem == "classification":
clusters = 1 if difficulty < 3 else 2
informs = 1 if difficulty < 4 else 2
data = sklearn.datasets.make_classification(
n_samples = number_samples,
n_features=number_features,
n_redundant=0,
class_sep=1/difficulty,
n_informative=informs,
n_clusters_per_class=clusters)
if problem == "blobs":
data = sklearn.datasets.make_blobs(
n_samples = number_samples,
n_features=number_features,
centers=number_classes,
cluster_std = difficulty)
if problem == "gaussian quantiles":
data = sklearn.datasets.make_gaussian_quantiles(mean=None,
cov=difficulty,
n_samples=number_samples,
n_features=number_features,
n_classes=number_classes,
shuffle=True,
random_state=None)
if problem == "moons":
data = sklearn.datasets.make_moons(
n_samples = number_samples)
if problem == "circles":
data = sklearn.datasets.make_circles(
n_samples = number_samples)
return data
data = load_data(problem)
X, Y = data
# Input Data
plt.figure("Input Data")
plt.scatter(X[:, 0], X[:, 1], c=Y, s=40, cmap=plt.cm.Spectral)2. 配置 NEAT
由于分类问题比 XOR 更复杂,因此需要修改 NEAT 配置文件 config 中的配置选项,最大适应度阈值为 3.0,增加一个中间节点层。实际上,在 NEAT 中,无需考虑节点的层次关系,相反,只关心输入/输出和中间节点的数量。如果这些中间节点碰巧对齐在同一层次中,只是一种巧合。最后,添加两个可能的函数 (identity 和 relu) 更新激活选项:
shell
[NEAT]
fitness_criterion = max
fitness_threshold = 3.0
pop_size = 250
reset_on_extinction = 1
[DefaultGenome]
num_inputs = 2
num_hidden = 10
num_outputs = 1
initial_connection = partial_direct 0.5
feed_forward = True
compatibility_disjoint_coefficient = 1.0
compatibility_weight_coefficient = 0.6
conn_add_prob = 0.2
conn_delete_prob = 0.2
node_add_prob = 0.2
node_delete_prob = 0.2
activation_default = sigmoid
activation_options = sigmoid identity relu
activation_mutate_rate = 0.1
aggregation_default = sum
aggregation_options = sum
aggregation_mutate_rate = 0.0
bias_init_mean = 0.0
bias_init_stdev = 1.0
bias_replace_rate = 0.1
bias_mutate_rate = 0.7
bias_mutate_power = 0.5
bias_max_value = 30.0
bias_min_value = -30.0
response_init_mean = 1.0
response_init_stdev = 0.0
response_replace_rate = 0.0
response_mutate_rate = 0.0
response_mutate_power = 0.0
response_max_value = 30.0
response_min_value = -30.0
weight_max_value = 30
weight_min_value = -30
weight_init_mean = 0.0
weight_init_stdev = 1.0
weight_mutate_rate = 0.8
weight_replace_rate = 0.1
weight_mutate_power = 0.5
enabled_default = True
enabled_mutate_rate = 0.01
[DefaultSpeciesSet]
compatibility_threshold = 3.0
[DefaultStagnation]
species_fitness_func = max
max_stagnation = 20
[DefaultReproduction]
elitism = 2
survival_threshold = 0.23. 使用 NEAT 解决分类问题
(1) 应用新配置选项的起始基因组网络的输出如下所示。需要注意的是,并非所有节点都连接到输出,节点 10 并未连接到输入或输出,这使得网络能够消除不需要的节点,从而避免过拟合或欠拟合:
python
config = neat.Config(neat.DefaultGenome, neat.DefaultReproduction,
neat.DefaultSpeciesSet, neat.DefaultStagnation,
'config')
print(config.genome_type, config.genome_config,config.pop_size)
key = "fred"
genome = config.genome_type(key)
genome.configure_new(config.genome_config)
net = neat.nn.FeedForwardNetwork.create(genome, config)
fitness = 4
for x, y in zip(X,Y):
output = net.activate(x)
fitness -= (output-y)**2
print(fitness)
import graphviz
def draw_net(config, genome, view=False, filename=None, node_names=None, show_disabled=True, prune_unused=False,
node_colors=None, fmt='svg'):
""" Receives a genome and draws a neural network with arbitrary topology. """
# Attributes for network nodes.
if graphviz is None:
print("This display is not available due to a missing optional dependency (graphviz)")
return
# If requested, use a copy of the genome which omits all components that won't affect the output.
if prune_unused:
genome = genome.get_pruned_copy(config.genome_config)
if node_names is None:
node_names = {}
assert type(node_names) is dict
if node_colors is None:
node_colors = {}
assert type(node_colors) is dict
node_attrs = {
'shape': 'circle',
'fontsize': '9',
'height': '0.2',
'width': '0.2'}
dot = graphviz.Digraph(format=fmt, node_attr=node_attrs)
inputs = set()
for k in config.genome_config.input_keys:
inputs.add(k)
name = node_names.get(k, str(k))
input_attrs = {'style': 'filled', 'shape': 'box', 'fillcolor': node_colors.get(k, 'lightgray')}
dot.node(name, _attributes=input_attrs)
outputs = set()
for k in config.genome_config.output_keys:
outputs.add(k)
name = node_names.get(k, str(k))
node_attrs = {'style': 'filled', 'fillcolor': node_colors.get(k, 'lightblue')}
dot.node(name, _attributes=node_attrs)
used_nodes = set(genome.nodes.keys())
for n in used_nodes:
if n in inputs or n in outputs:
continue
attrs = {'style': 'filled',
'fillcolor': node_colors.get(n, 'white')}
dot.node(str(n), _attributes=attrs)
for cg in genome.connections.values():
if cg.enabled or show_disabled:
# if cg.input not in used_nodes or cg.output not in used_nodes:
# continue
input, output = cg.key
a = node_names.get(input, str(input))
b = node_names.get(output, str(output))
style = 'solid' if cg.enabled else 'dotted'
color = 'green' if cg.weight > 0 else 'red'
width = str(0.1 + abs(cg.weight / 5.0))
dot.edge(a, b, _attributes={'style': style, 'color': color, 'penwidth': width})
dot.render(filename, view=view)
dot.view()
return dot
node_names = {-1: 'X1', -2: 'X2', 0: 'Output'}
draw_net(config, genome, True, node_names=node_names)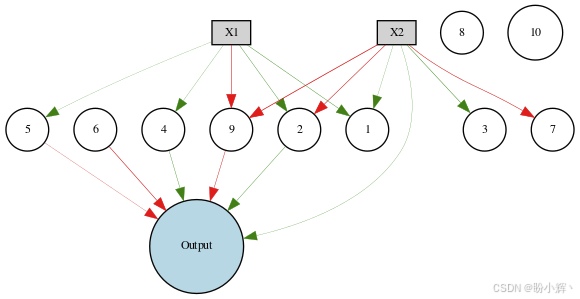
(2) 定义 show_predictions() 函数,调用 net.activate() 函数:
python
def eval_genomes(genomes, config):
for genome_id, genome in genomes:
genome.fitness = 4.0
net = neat.nn.FeedForwardNetwork.create(genome, config)
for x, y in zip(X,Y):
output = net.activate(x)[0]
genome.fitness -= (output-y)**2
def show_predictions(net, X, Y, name=""):
""" display the labeled data X and a surface of prediction of model """
x_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1
y_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, 0.01), np.arange(y_min, y_max, 0.01))
X_temp = np.c_[xx.flatten(), yy.flatten()]
Z = []
print(X_temp.shape)
for x in X_temp:
Z.append(net.activate(x))
Z = np.array(Z)
plt.figure("Predictions " + name)
plt.contourf(xx, yy, Z.reshape(xx.shape), cmap=plt.cm.Spectral)
plt.ylabel('x2')
plt.xlabel('x1')
plt.scatter(X[:, 0], X[:, 1],c=Y, s=40, cmap=plt.cm.Spectral)
plt.show()
show_predictions(net, X, Y)(3) 定义 CustomReporter 类,NEAT-Python 允许进行各种自定义,添加用于可视化拟合进展的代码。在 CustomReporter 类中,在评估基因组后的 post_evaluate() 函数中添加了自定义代码。我们不希望在每次迭代时都渲染,因此添加模数检查,由 init 函数中设置的 self.gen_display 参数控制。需要渲染时,从基因组创建网络,并在 update_show_predictions 函数中对其进行评估:
python
from neat.math_util import mean, stdev
class CustomReporter(neat.reporting.BaseReporter):
"""Uses `print` to output information about the run; an example reporter class."""
def __init__(self, show_species_detail, gen_display=100):
self.show_species_detail = show_species_detail
self.generation = None
self.generation_start_time = None
self.generation_times = []
self.num_extinctions = 0
self.gen_display = gen_display
def start_generation(self, generation):
clear_output()
self.generation = generation
print('\n ****** Running generation {0} ****** \n'.format(generation))
self.generation_start_time = time.time()
def end_generation(self, config, population, species_set):
ng = len(population)
ns = len(species_set.species)
if self.show_species_detail:
print('Population of {0:d} members in {1:d} species:'.format(ng, ns))
print(" ID age size fitness adj fit stag")
print(" ==== === ==== ========= ======= ====")
for sid in sorted(species_set.species):
s = species_set.species[sid]
a = self.generation - s.created
n = len(s.members)
f = "--" if s.fitness is None else f"{s.fitness:.3f}"
af = "--" if s.adjusted_fitness is None else f"{s.adjusted_fitness:.3f}"
st = self.generation - s.last_improved
print(f" {sid:>4} {a:>3} {n:>4} {f:>9} {af:>7} {st:>4}")
else:
print('Population of {0:d} members in {1:d} species'.format(ng, ns))
elapsed = time.time() - self.generation_start_time
self.generation_times.append(elapsed)
self.generation_times = self.generation_times[-10:]
average = sum(self.generation_times) / len(self.generation_times)
print('Total extinctions: {0:d}'.format(self.num_extinctions))
if len(self.generation_times) > 1:
print("Generation time: {0:.3f} sec ({1:.3f} average)".format(elapsed, average))
else:
print("Generation time: {0:.3f} sec".format(elapsed))
def post_evaluate(self, config, population, species, best_genome):
# pylint: disable=no-self-use
fitnesses = [c.fitness for c in population.values()]
fit_mean = mean(fitnesses)
fit_std = stdev(fitnesses)
best_species_id = species.get_species_id(best_genome.key)
print('Population\'s average fitness: {0:3.5f} stdev: {1:3.5f}'.format(fit_mean, fit_std))
print(
'Best fitness: {0:3.5f} - size: {1!r} - species {2} - id {3}'.format(best_genome.fitness,
best_genome.size(),
best_species_id,
best_genome.key))
if (self.generation) % self.gen_display == 0 :
net = neat.nn.FeedForwardNetwork.create(best_genome, config)
show_predictions(net, X, Y)
time.sleep(5)
def complete_extinction(self):
self.num_extinctions += 1
print('All species extinct.')
def found_solution(self, config, generation, best):
print('\nBest individual in generation {0} meets fitness threshold - complexity: {1!r}'.format(
self.generation, best.size()))
def species_stagnant(self, sid, species):
if self.show_species_detail:
print("\nSpecies {0} with {1} members is stagnated: removing it".format(sid, len(species.members)))
def info(self, msg):
print(msg)(4) 向种群 p 添加 CustomReporter 的代码,使用 add_reporter 函数调用,该自定义 Reporter 允许我们微调进化输出,并能够添加可视化:
python
DISPLAY_ON_GENERATION = 100 #@param {type:"slider", min:1, max:100, step:1}
# Create the population, which is the top-level object for a NEAT run.
p = neat.Population(config)
# Add a stdout reporter to show progress in the terminal.
p.add_reporter(CustomReporter(False, gen_display=DISPLAY_ON_GENERATION))
# Run until a solution is found.
winner = p.run(eval_genomes)
# Display the winning genome.
print('\nBest genome:\n{!s}'.format(winner))
# Show output of the most fit genome against training data.
print('\nOutput:')
winner_net = neat.nn.FeedForwardNetwork.create(winner, config)
show_predictions(winner_net, X, Y)
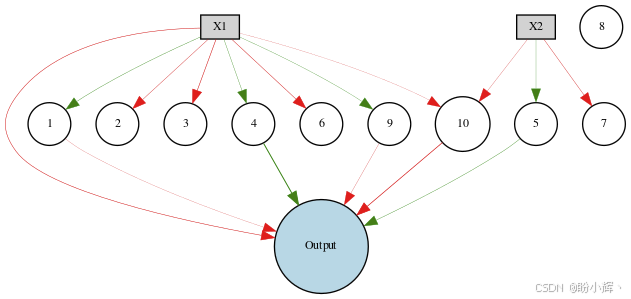
draw_net(config, winner, True, node_names=node_names)在本节所用数据集上进化 NEAT 网络的结果如下所示,可以看到,大多数网络节点未连接到输出节点。
小结
使用 NEAT 解决分类问题通过进化的方式自动优化神经网络的结构和权重。结合适当的数据预处理和参数设置,可以有效地提高分类任务的性能。在本节中,我们使用 sklearn 库构建示例数据集,利用 NEAT 解决分类问题并可视化 NEAT 分类结果。
系列链接
遗传算法与深度学习实战(1)------进化深度学习
遗传算法与深度学习实战(2)------生命模拟及其应用
遗传算法与深度学习实战(3)------生命模拟与进化论
遗传算法与深度学习实战(4)------遗传算法(Genetic Algorithm)详解与实现
遗传算法与深度学习实战(5)------遗传算法中常用遗传算子
遗传算法与深度学习实战(6)------遗传算法框架DEAP
遗传算法与深度学习实战(7)------DEAP框架初体验
遗传算法与深度学习实战(8)------使用遗传算法解决N皇后问题
遗传算法与深度学习实战(9)------使用遗传算法解决旅行商问题
遗传算法与深度学习实战(10)------使用遗传算法重建图像
遗传算法与深度学习实战(11)------遗传编程详解与实现
遗传算法与深度学习实战(12)------粒子群优化详解与实现
遗传算法与深度学习实战(13)------协同进化详解与实现
遗传算法与深度学习实战(14)------进化策略详解与实现
遗传算法与深度学习实战(15)------差分进化详解与实现
遗传算法与深度学习实战(16)------神经网络超参数优化
遗传算法与深度学习实战(17)------使用随机搜索自动超参数优化
遗传算法与深度学习实战(18)------使用网格搜索自动超参数优化
遗传算法与深度学习实战(19)------使用粒子群优化自动超参数优化
遗传算法与深度学习实战(20)------使用进化策略自动超参数优化
遗传算法与深度学习实战(21)------使用差分搜索自动超参数优化
遗传算法与深度学习实战(22)------使用Numpy构建神经网络
遗传算法与深度学习实战(23)------利用遗传算法优化深度学习模型
遗传算法与深度学习实战(24)------在Keras中应用神经进化优化
遗传算法与深度学习实战(25)------使用Keras构建卷积神经网络
遗传算法与深度学习实战(26)------编码卷积神经网络架构
遗传算法与深度学习实战(27)------进化卷积神经网络
遗传算法与深度学习实战(28)------卷积自编码器详解与实现
遗传算法与深度学习实战(29)------编码卷积自编码器架构
遗传算法与深度学习实战(30)------使用遗传算法优化自编码器模型
遗传算法与深度学习实战(31)------变分自编码器详解与实现
遗传算法与深度学习实战(32)------生成对抗网络详解与实现
遗传算法与深度学习实战(33)------WGAN详解与实现
遗传算法与深度学习实战(34)------编码WGAN
遗传算法与深度学习实战(35)------使用遗传算法优化生成对抗网络
遗传算法与深度学习实战(36)------NEAT详解与实现