Clustering
介绍
分组变量将样本的观测值划分为可以相互比较属性的组。例如,观测值的分组可以是聚类算法的结果或手动空间分割的结果。本教程将展示如何在SPATA2中应用和添加聚类。
go
# load required packages
library(SPATA2)
library(SPATAData)
library(tidyverse)
go
object_t269<- readRDS("object_t269.rds")
object_t269 <- updateSpataObject(object_t269)
go
# plot histology
plotSurface(object_t269, color_by = "histology", pt_clrp = "npg")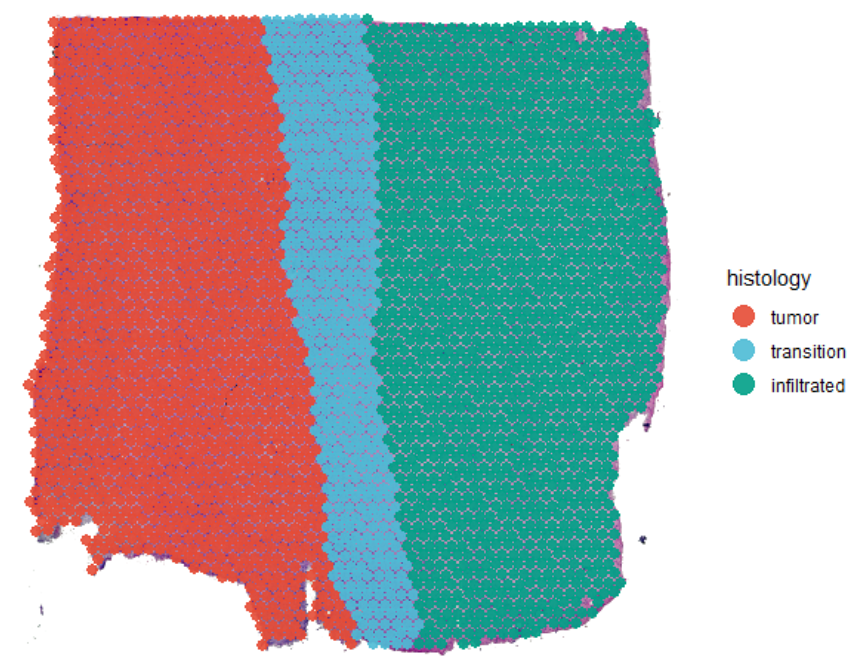
go
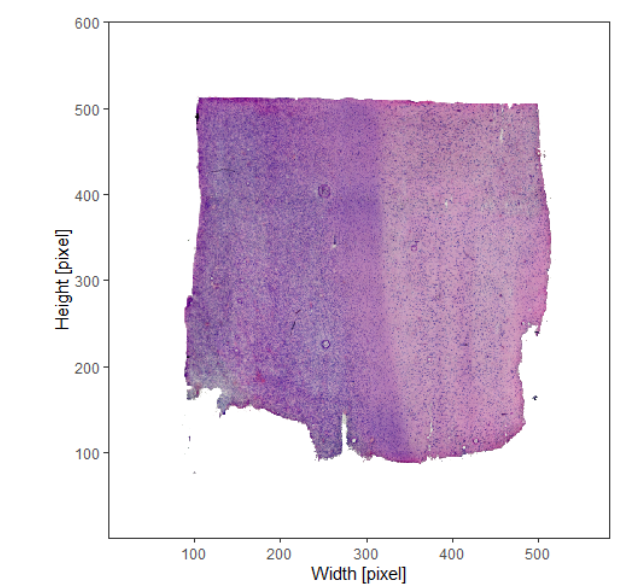
plotImage(object_t269)2. SPATA2 内的聚类
有许多算法可以将您的样本分成子组。SPATA2 提供了多种聚类算法的封装。那些会立即将结果添加到 SPATA2 对象中的聚类算法,其名称以 run-* 开头,并以 *-Clustering() 结尾。例如:runBayesSpaceClustering()、runKmeansClustering()、runSeuratClustering()。参数名称或命名指定了输出分组变量的名称。可通过getGroupingOptions()获取结果分组变量名称。
go
# current grouping options
getGroupingOptions(object_t269)
go
## factor factor factor factor
## "tissue_section" "seurat_clusters" "histology" "bayes_space"
go
# run the pipeline
object_t269 <-
runBayesSpaceClustering(
object = object_t269,
name = "bayes_spacev2", # the name of the output grouping variable
qs = 5
)
go
# run PCA based on which clustering is conducted
object_t269 <- runPCA(object_t269, n_pcs = 20)
object_t269 <-
runKmeansClustering(
object = object_t269,
ks = c(7, 8),
methods_kmeans = "Lloyd"
)
go
# results are immediately stored in the objects feature data
getGroupingOptions(object_t269)
go
## factor factor factor factor
## "tissue_section" "seurat_clusters" "histology" "bayes_space"
## factor factor factor
## "bayes_spacev2" "Lloyd_k7" "Lloyd_k8"
go
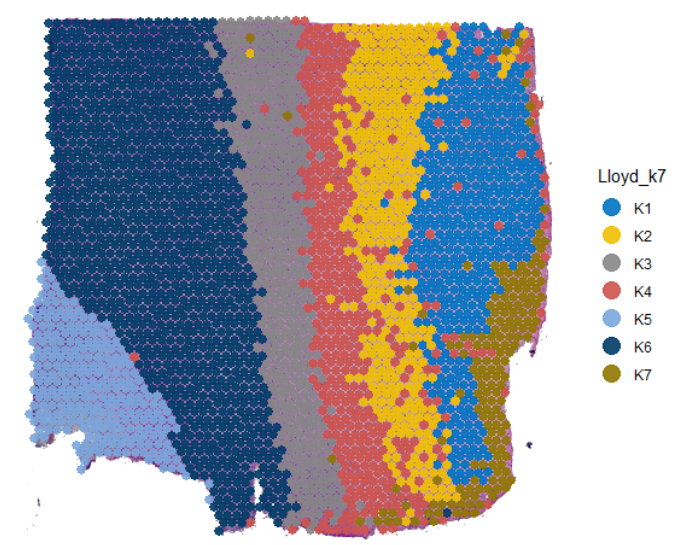
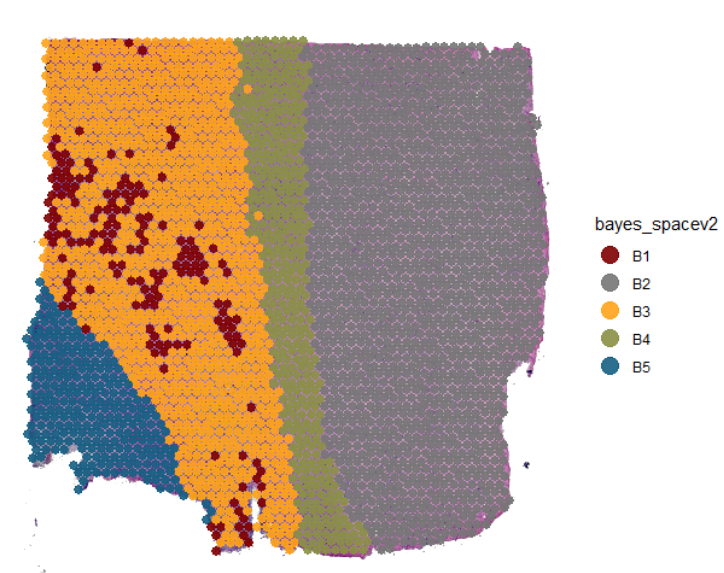
# left plot
plotSurface(
object = object_t269,
color_by = "bayes_spacev2",
pt_clrp = "uc"
)
go
# right plot
plotSurface(
object = object_t269,
color_by = "Lloyd_k7",
pt_clrp = "jco"
)3. SPATA2以外的聚类
聚类可能由多种聚类算法产生。如果这些算法未在SPATA2函数中实现,可以使用addFeatures()函数将它们添加进去。唯一的要求是有一个名为barcodes的变量,用于将组映射到观测值。请注意,变量必须为因子类(factor class),才能被识别为分组变量。
go
# uses kmeans outside of SPATA2
kmeans_res <-
stats::kmeans(
x = getPcaMtr(object_t269),
centers = 7,
algorithm = "Hartigan-Wong"
)
head(kmeans_res[["cluster"]])
go
## GTAGCGCTGTTGTAGT-1 TTGTTTGTGTAAATTC-1 CGTAGCGCCGACGTTG-1 GTAGACAACCGATGAA-1
## 2 4 4 4
## ACAGATTAGGTTAGTG-1 TGAGATCAAATACTCA-1
## 2 2
go
cluster_df <-
as.data.frame(kmeans_res[["cluster"]]) %>%
tibble::rownames_to_column(var = "barcodes") %>%
magrittr::set_colnames(value = c("barcodes", "kmeans_4_HW")) %>%
tibble::as_tibble()
cluster_df[["kmeans_4_HW"]] <- as.factor(cluster_df[["kmeans_4_HW"]])
cluster_df
go
## # A tibble: 3,213 × 2
## barcodes kmeans_4_HW
## <chr> <fct>
## 1 GTAGCGCTGTTGTAGT-1 2
## 2 TTGTTTGTGTAAATTC-1 4
## 3 CGTAGCGCCGACGTTG-1 4
## 4 GTAGACAACCGATGAA-1 4
## 5 ACAGATTAGGTTAGTG-1 2
## 6 TGAGATCAAATACTCA-1 2
## 7 CTGGTCCTAACTTGGC-1 2
## 8 TGCACGAGTCGGCAGC-1 3
## 9 ATAGTCTTTGACGTGC-1 2
## 10 GGGTGGTCCAGCCTGT-1 3
## # ℹ 3,203 more rows
go
# grouping options before adding
getGroupingOptions(object_t269)
go
## factor factor factor factor
## "tissue_section" "seurat_clusters" "histology" "bayes_space"
## factor factor factor
## "bayes_spacev2" "Lloyd_k7" "Lloyd_k8"
go
# add the cluster results to the meta features
object_t269 <-
addFeatures(
object = object_t269,
feature_df = cluster_df
)
# grouping options names afterwards
getGroupingOptions(object_t269)
go
## factor factor factor factor
## "tissue_section" "seurat_clusters" "histology" "bayes_space"
## factor factor factor factor
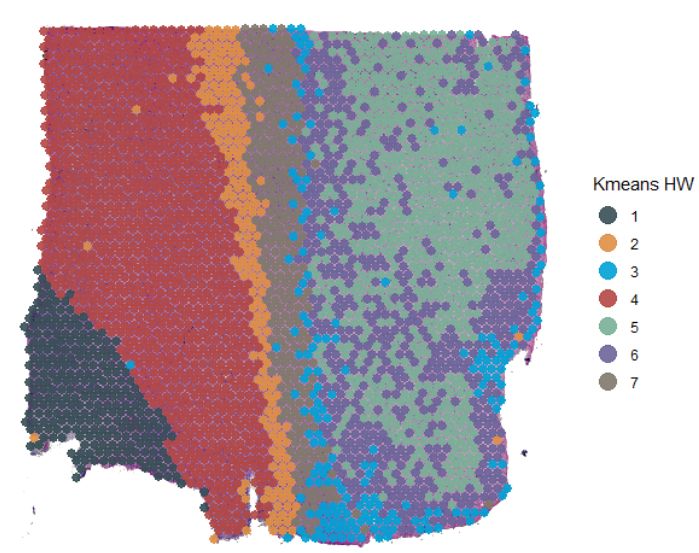
## "bayes_spacev2" "Lloyd_k7" "Lloyd_k8" "kmeans_4_HW"继续通过可视化结果或使用差异表达分析(DEA)来研究其转录特征。
go
plotSurface(
object = object_t269,
color_by = "kmeans_4_HW",
pt_clrp = "jama"
) +
labs(color = "Kmeans HW")