加载模块
import math
import itertools
import numpy as np
import pandas as pd
import seaborn as sns
import tensorflow as tf
from keras import layers
from sklearn.svm import SVR
from tensorflow import keras
from keras import backend as K
import matplotlib.pyplot as plt
from keras.regularizers import l2
from keras.regularizers import l1_l2
from keras.models import load_model
from keras.callbacks import ReduceLROnPlateau
from sklearn.model_selection import KFold, GridSearchCV
from sklearn.preprocessing import StandardScaler
from sklearn.pipeline import make_pipeline
from sklearn.model_selection import GroupShuffleSplit
from sklearn.metrics import mean_squared_error, r2_score
from sklearn.model_selection import train_test_split, cross_val_score, cross_validate
from keras.callbacks import Callback, EarlyStopping, ModelCheckpoint, TensorBoard, LambdaCallback
from keras.layers import Input, Dense, Lambda, LSTM, RepeatVector, Bidirectional, Masking, Dropout, BatchNormalization读取数据
# Data reading
data_dir = './original_data/'
# Specify the file path and column names
file_path = os.path.join(data_dir, 'battery_RUL.txt')
# Specify the column names
column_names = [
'unit_nr',
'time_cycles',
's_discharge_t',
's_decrement_3.6-3.4V',
's_max_voltage_discharge',
's_min_voltage_charge',
'Time_at_4.15V_s',
's_time_constant_current',
's_charging_time',
'RUL'
]
# Read the text file into a DataFrame
df = pd.read_csv(file_path, sep='\s+', header=None, skiprows=0, names=column_names)
# Calculate the unique number of batteries
unique_batteries = df['unit_nr'].nunique()
print("\n Unique number of batteries:", unique_batteries)
# Check for missing values
if df.isnull().any().any():
print("\n There are missing values in the DataFrame.")
# Print the shape of the DataFrame
print("\n DataFrame shape:", df.shape)
# print df info
print(df.info())
# Print the first few rows of the DataFrame
print("\n First few rows of the DataFrame:")
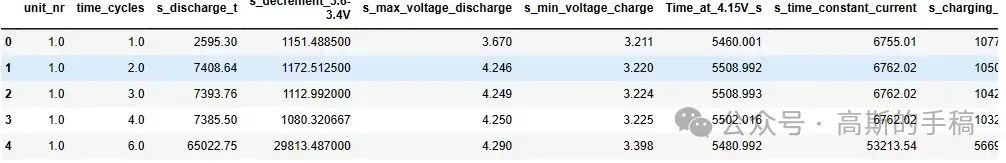
df.head()Unique number of batteries: 14
DataFrame shape: (15064, 10)
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 15064 entries, 0 to 15063
Data columns (total 10 columns):
Column Non-Null Count Dtype
0 unit_nr 15064 non-null float64
1 time_cycles 15064 non-null float64
2 s_discharge_t 15064 non-null float64
3 s_decrement_3.6-3.4V 15064 non-null float64
4 s_max_voltage_discharge 15064 non-null float64
5 s_min_voltage_charge 15064 non-null float64
6 Time_at_4.15V_s 15064 non-null float64
7 s_time_constant_current 15064 non-null float64
8 s_charging_time 15064 non-null float64
9 RUL 15064 non-null int64
dtypes: float64(9), int64(1)
memory usage: 1.1 MB
None
First few rows of the DataFrame:
数据前处理
# Filter rows where 'time_cycles' is equal to 1
df_at_cycle_1 = df[df['time_cycles'] == 1]
# Create a bar chart with all battery units on the X-axis
plt.figure(figsize=(12, 6))
plt.bar(df_at_cycle_1['unit_nr'], df_at_cycle_1['RUL'])
plt.xlabel('Battery Number')
plt.ylabel('RUL at Cycle 1')
plt.title('RUL Values at the Beginning of Cycle 1 for Each Battery Unit')
plt.xticks(df_at_cycle_1['unit_nr']) # Set X-axis ticks explicitly
plt.tight_layout()
plt.show()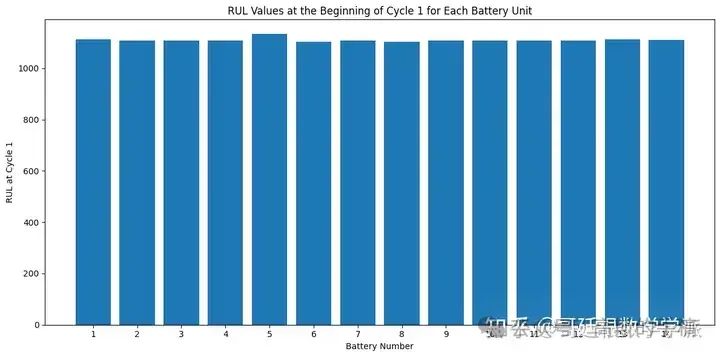
plt.figure(figsize = (8,8))
sns.heatmap(df.corr(),annot=True, cbar=False, cmap='Blues', fmt='.1f')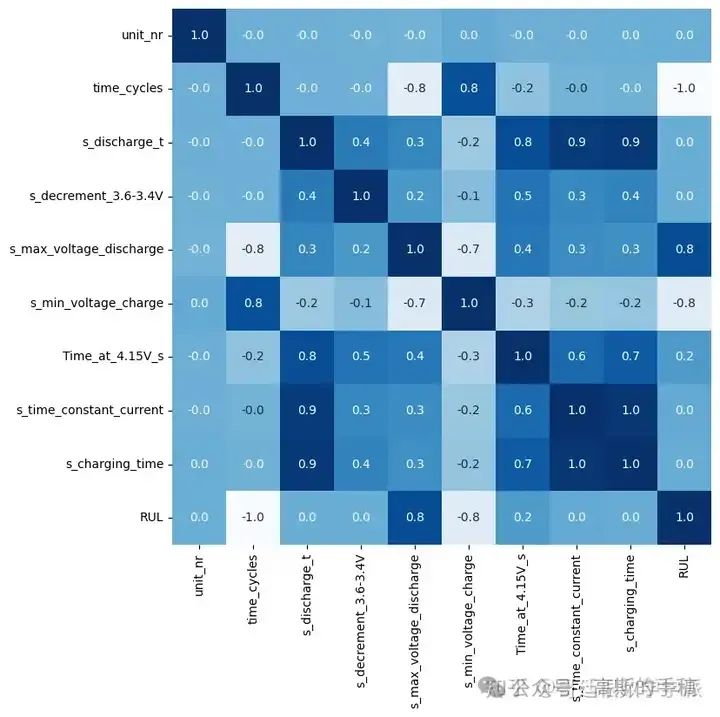
Correlation between RUL and:
Max. Voltage Dischar. (V) is 0.8
Min. Voltage Charg. (V) is -0.8
Time at 4.15V (s) is 0.2
Cycle index is -1.0
Discharge Time (s), Decrement 3.6-3.4V (s), Time constant current (s) and Charging time (s) are 0.
And correlation between Time at 4.15V and these four features are 0.8, 0.5,0.6 and 0.7.
df1=df.drop(['s_discharge_t', 's_decrement_3.6-3.4V', 's_time_constant_current','s_charging_time'], axis=1)
plt.figure(figsize = (4,4))
sns.heatmap(df1.corr(),annot=True, cbar=False, cmap='Blues', fmt='.1f')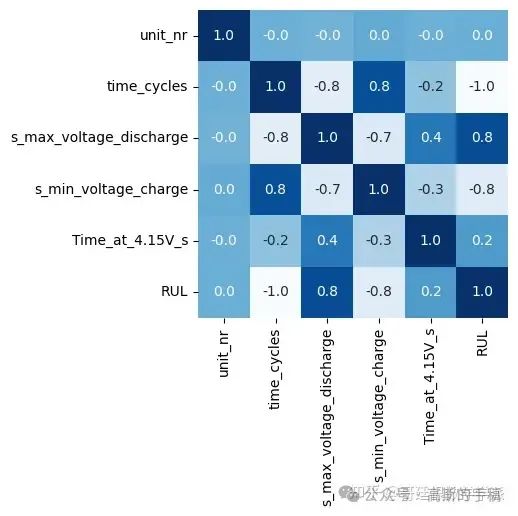
df1.head()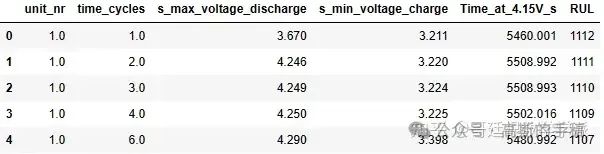
def exponential_smoothing(df, sensors, n_samples, alpha=0.4):
df = df.copy()
# first, take the exponential weighted mean
df[sensors] = df.groupby('unit_nr', group_keys=True)[sensors].apply(lambda x: x.ewm(alpha=alpha).mean()).reset_index(level=0, drop=True)
# second, drop first n_samples of each unit_nr to reduce filter delay
def create_mask(data, samples):
result = np.ones_like(data)
result[0:samples] = 0
return result
mask = df.groupby('unit_nr')['unit_nr'].transform(create_mask, samples=n_samples).astype(bool)
df = df[mask]
return df
#-----------------------------------------------------------------------------------------------------------------------
def data_standardization(df, sensors):
df = df.copy()
# Apply StandardScaler to the sensor data
scaler = StandardScaler()
df[sensors] = scaler.fit_transform(df[sensors])
return df
# MMS_X = MinMaxScaler()
# mms_y = MinMaxScaler()
#-----------------------------------------------------------------------------------------------------------------------
def gen_train_data(df, sequence_length, columns):
data = df[columns].values
num_elements = data.shape[0]
# -1 and +1 because of Python indexing
for start, stop in zip(range(0, num_elements-(sequence_length-1)), range(sequence_length, num_elements+1)):
yield data[start:stop, :]
#-----------------------------------------------------------------------------------------------------------------------
def gen_data_wrapper(df, sequence_length, columns, unit_nrs=np.array([])):
if unit_nrs.size <= 0:
unit_nrs = df['unit_nr'].unique()
data_gen = (list(gen_train_data(df[df['unit_nr']==unit_nr], sequence_length, columns))
for unit_nr in unit_nrs)
data_array = np.concatenate(list(data_gen)).astype(np.float32)
return data_array
#-----------------------------------------------------------------------------------------------------------------------
def gen_labels(df, sequence_length, label):
data_matrix = df[label].values
num_elements = data_matrix.shape[0]
# -1 because I want to predict the rul of that last row in the sequence, not the next row
return data_matrix[sequence_length-1:num_elements, :]
#-----------------------------------------------------------------------------------------------------------------------
def gen_label_wrapper(df, sequence_length, label, unit_nrs=np.array([])):
if unit_nrs.size <= 0:
unit_nrs = df['unit_nr'].unique()
label_gen = [gen_labels(df[df['unit_nr']==unit_nr], sequence_length, label)
for unit_nr in unit_nrs]
label_array = np.concatenate(label_gen).astype(np.float32)
return label_array
#---------Original code------------------------------------------------------------------------------------------------------
def gen_test_data(df, sequence_length, columns, mask_value):
if df.shape[0] < sequence_length:
data_matrix = np.full(shape=(sequence_length, len(columns)), fill_value=mask_value) # pad
idx = data_matrix.shape[0] - df.shape[0]
data_matrix[idx:,:] = df[columns].values # fill with available data
else:
data_matrix = df[columns].values
# specifically yield the last possible sequence
stop = data_matrix.shape[0]
start = stop - sequence_length
for i in list(range(1)):
yield data_matrix[start:stop, :]
#---------------------------------------------------------------------------------------------------------------------------
def change_test_index(df, initial_unit_number, start_index, end_index, min_rows, max_rows):
df.reset_index(drop=True, inplace=True)
y_test = [] # Initialize an empty list to store y_test values
y_test.append(df.loc[0, 'RUL'])
while end_index < len(df):
# Calculate the number of rows to be assigned to the current unit_nr
num_rows = min_rows + (end_index % (max_rows - min_rows + 1))
end_index = end_index + num_rows
# Update the unit_nr for the current block of rows
df.loc[start_index:end_index, 'unit_nr'] = initial_unit_number
# Update the "time_cycles" column starting from the first row with "time_cycles" == 1
time_cycle_to_change = end_index + 1
df.loc[time_cycle_to_change, 'time_cycles'] = 1
# Append the values to y_test (replace 'column_name' with the actual column name you want to append)
y_test.append(df.loc[time_cycle_to_change, 'RUL'])
# Update the starting and ending index for the next block of rows
start_index = end_index + 1
initial_unit_number += 1
# Drop rows with NaN values at the end of DataFrame 'df'
df.dropna(axis=0, how='any', inplace=True)
# Drop any NaN values at the end of list 'y_test'
while len(y_test) > 0 and pd.isnull(y_test[-1]):
y_test.pop()
return df, y_test获取数据
def get_data(df, sensors, sequence_length, alpha):
# List of battery units
battery_units = range(1, 15)
sensor_names = ['s_{}'.format(i+1) for i in range(0,10)]
# Define the number of batteries for training and testing
train_batteries = 12
test_batteries = 2
# Extract the batteries for training and testing
train_units = battery_units[:train_batteries]
test_units = battery_units[train_batteries:train_batteries + test_batteries]
# Create the training and testing datasets
train = df[df['unit_nr'].isin(train_units)].copy() # Use copy to avoid the SettingWithCopyWarning
test = df[df['unit_nr'].isin(test_units)].copy() # Use copy to avoid the SettingWithCopyWarning
X_test_pre, y_test = change_test_index(test, 13, 0, 0, 230, 240)
# y_test = pd.Series(y_test) # Convert y_test list to a pandas Series
# remove unused sensors
drop_sensors = [element for element in sensor_names if element not in sensors]
# Apply standardization to the training and testing data using data_standardization function
standard_train = data_standardization(train, sensors)
standard_test = data_standardization(test, sensors)
# Exponential smoothing of training and testing data
X_train_pre = exponential_smoothing(standard_train, sensors, 0, alpha)
X_test_pre = exponential_smoothing(standard_test, sensors, 0, alpha)
# Train-validation split
gss = GroupShuffleSplit(n_splits=1, train_size=0.85, random_state=42)
# Generate the train/val for each sample by iterating over the train and val units
for train_unit, val_unit in gss.split(X_train_pre['unit_nr'].unique(), groups=X_train_pre['unit_nr'].unique()):
train_unit = X_train_pre['unit_nr'].unique()[train_unit] # gss returns indexes and index starts at 1
val_unit = X_train_pre['unit_nr'].unique()[val_unit]
x_train = gen_data_wrapper(X_train_pre, sequence_length, sensors, train_unit)
y_train = gen_label_wrapper(X_train_pre, sequence_length, ['RUL'], train_unit)
x_val = gen_data_wrapper(X_train_pre, sequence_length, sensors, val_unit)
y_val = gen_label_wrapper(X_train_pre, sequence_length, ['RUL'], val_unit)
# create sequences for test
test_gen = (list(gen_test_data(X_test_pre[X_test_pre['unit_nr']==unit_nr], sequence_length, sensors, -99.))
for unit_nr in X_test_pre['unit_nr'].unique())
x_test = np.concatenate(list(test_gen)).astype(np.float32)
return x_train, y_train, x_val, y_val, x_test, y_test训练回调
# --------------------------------------- TRAINING CALLBACKS ---------------------------------------
class save_latent_space_viz(Callback):
def __init__(self, model, data, target):
self.model = model
self.data = data
self.target = target
def on_train_begin(self, logs={}):
self.best_val_loss = 100000
def on_epoch_end(self, epoch, logs=None):
encoder = self.model.layers[0]
if logs.get('val_loss') < self.best_val_loss:
self.best_val_loss = logs.get('val_loss')
viz_latent_space(encoder, self.data, self.target, epoch, True, False)
def get_callbacks(model, data, target):
model_callbacks = [
EarlyStopping(monitor='val_loss', mode='min', verbose=1, patience=30),
ModelCheckpoint(filepath='./checkpoints/checkpoint',monitor='val_loss', mode='min', verbose=1, save_best_only=True, save_weights_only=True),
TensorBoard(log_dir='./logs'),
save_latent_space_viz(model, data, target)
]
return model_callbacks
def viz_latent_space(encoder, data, targets=[], epoch='Final', save=False, show=True):
z, _, _ = encoder.predict(data)
plt.figure(figsize=(3, 3)) # Smaller figsize value to reduce the plot size
if len(targets) > 0:
plt.scatter(z[:, 1], z[:, 0], c=targets)
else:
plt.scatter(z[:, 1], z[:, 0])
plt.xlabel('z - dim 1')
plt.ylabel('z - dim 2')
plt.colorbar()
if show:
plt.show()
if save:
plt.savefig('./images/latent_space_epoch' + str(epoch) + '.png')
return z最佳学习率
# ----------------------------------------- FIND OPTIMAL LR ----------------------------------------
class LRFinder:
"""
Cyclical LR, code tailored from:
https://towardsdatascience.com/estimating-optimal-learning-rate-for-a-deep-neural-network-ce32f2556ce0
"""
def __init__(self, model):
self.model = model
self.losses = []
self.lrs = []
self.best_loss = 1e9
def on_batch_end(self, batch, logs):
# Log the learning rate
lr = K.get_value(self.model.optimizer.lr)
self.lrs.append(lr)
# Log the loss
loss = logs['loss']
self.losses.append(loss)
# Check whether the loss got too large or NaN
if batch > 5 and (math.isnan(loss) or loss > self.best_loss * 4):
self.model.stop_training = True
return
if loss < self.best_loss:
self.best_loss = loss
# Increase the learning rate for the next batch
lr *= self.lr_mult
K.set_value(self.model.optimizer.lr, lr)
def find(self, x_train, y_train, start_lr, end_lr, batch_size=64, epochs=1, **kw_fit):
# If x_train contains data for multiple inputs, use length of the first input.
# Assumption: the first element in the list is single input; NOT a list of inputs.
N = x_train[0].shape[0] if isinstance(x_train, list) else x_train.shape[0]
# Compute number of batches and LR multiplier
num_batches = epochs * N / batch_size
self.lr_mult = (float(end_lr) / float(start_lr)) ** (float(1) / float(num_batches))
# Save weights into a file
initial_weights = self.model.get_weights()
# Remember the original learning rate
original_lr = K.get_value(self.model.optimizer.lr)
# Set the initial learning rate
K.set_value(self.model.optimizer.lr, start_lr)
callback = LambdaCallback(on_batch_end=lambda batch, logs: self.on_batch_end(batch, logs))
self.model.fit(x_train, y_train,
batch_size=batch_size, epochs=epochs,
callbacks=[callback],
**kw_fit)
# Restore the weights to the state before model fitting
self.model.set_weights(initial_weights)
# Restore the original learning rate
K.set_value(self.model.optimizer.lr, original_lr)
def find_generator(self, generator, start_lr, end_lr, epochs=1, steps_per_epoch=None, **kw_fit):
if steps_per_epoch is None:
try:
steps_per_epoch = len(generator)
except (ValueError, NotImplementedError) as e:
raise e('`steps_per_epoch=None` is only valid for a'
' generator based on the '
'`keras.utils.Sequence`'
' class. Please specify `steps_per_epoch` '
'or use the `keras.utils.Sequence` class.')
self.lr_mult = (float(end_lr) / float(start_lr)) ** (float(1) / float(epochs * steps_per_epoch))
# Save weights into a file
initial_weights = self.model.get_weights()
# Remember the original learning rate
original_lr = K.get_value(self.model.optimizer.lr)
# Set the initial learning rate
K.set_value(self.model.optimizer.lr, start_lr)
callback = LambdaCallback(on_batch_end=lambda batch,
logs: self.on_batch_end(batch, logs))
self.model.fit_generator(generator=generator,
epochs=epochs,
steps_per_epoch=steps_per_epoch,
callbacks=[callback],
**kw_fit)
# Restore the weights to the state before model fitting
self.model.set_weights(initial_weights)
# Restore the original learning rate
K.set_value(self.model.optimizer.lr, original_lr)
def plot_loss(self, n_skip_beginning=10, n_skip_end=5, x_scale='log'):
"""
Plots the loss.
Parameters:
n_skip_beginning - number of batches to skip on the left.
n_skip_end - number of batches to skip on the right.
"""
plt.ylabel("loss")
plt.xlabel("learning rate (log scale)")
plt.plot(self.lrs[n_skip_beginning:-n_skip_end], self.losses[n_skip_beginning:-n_skip_end])
plt.xscale(x_scale)
plt.show()
def plot_loss_change(self, sma=1, n_skip_beginning=10, n_skip_end=5, y_lim=(-0.01, 0.01)):
"""
Plots rate of change of the loss function.
Parameters:
sma - number of batches for simple moving average to smooth out the curve.
n_skip_beginning - number of batches to skip on the left.
n_skip_end - number of batches to skip on the right.
y_lim - limits for the y axis.
"""
derivatives = self.get_derivatives(sma)[n_skip_beginning:-n_skip_end]
lrs = self.lrs[n_skip_beginning:-n_skip_end]
plt.ylabel("rate of loss change")
plt.xlabel("learning rate (log scale)")
plt.plot(lrs, derivatives)
plt.xscale('log')
plt.ylim(y_lim)
plt.show()
def get_derivatives(self, sma):
assert sma >= 1
derivatives = [0] * sma
for i in range(sma, len(self.lrs)):
derivatives.append((self.losses[i] - self.losses[i - sma]) / sma)
return derivatives
def get_best_lr(self, sma, n_skip_beginning=10, n_skip_end=5):
derivatives = self.get_derivatives(sma)
best_der_idx = np.argmin(derivatives[n_skip_beginning:-n_skip_end])
return self.lrs[n_skip_beginning:-n_skip_end][best_der_idx]结果
# --------------------------------------------- RESULTS --------------------------------------------
def get_model(path):
saved_VRAE_model = load_model(path, compile=False)
# return encoder, regressor
return saved_VRAE_model.layers[1], saved_VRAE_model.layers[2]
def evaluate(y_true, y_hat, label='test'):
mse = mean_squared_error(y_true, y_hat)
rmse = np.sqrt(mse)
variance = r2_score(y_true, y_hat)
print('{} set RMSE:{}, R2:{}'.format(label, rmse, variance))
return rmse, variance
def score(y_true, y_hat):
res = 0
for true, hat in zip(y_true, y_hat):
subs = hat - true
if subs < 0:
res = res + np.exp(-subs/10)[0]-1
else:
res = res + np.exp(subs/13)[0]-1
print("score: ", res)
def results(path, x_train, y_train, x_test, y_test):
# Get model
encoder, regressor = get_model(path)
# Latent space
train_mu = viz_latent_space(encoder, x_train, y_train)
test_mu = viz_latent_space(encoder, x_test, y_test)
# Evaluate
y_hat_train = regressor.predict(train_mu)
y_hat_test = regressor.predict(test_mu)
evaluate(y_train, y_hat_train, 'train')
evaluate(y_test, y_hat_test, 'test')
score(y_test, y_hat_test)模型结构
class Sampling(keras.layers.Layer):
"""Uses (z_mean, sigma) to sample z, the vector encoding an engine trajetory."""
def call(self, inputs):
mu, sigma = inputs
batch = tf.shape(mu)[0]
dim = tf.shape(mu)[1]
epsilon = tf.keras.backend.random_normal(shape=(batch, dim))
return mu + tf.exp(0.5 * sigma) * epsilon
class RVE(keras.Model):
def __init__(self, encoder, regressor, decoder=None, **kwargs):
super(RVE, self).__init__(**kwargs)
self.encoder = encoder
self.regressor = regressor
self.total_loss_tracker = keras.metrics.Mean(name="total_loss")
self.kl_loss_tracker = keras.metrics.Mean(name="kl_loss")
self.reg_loss_tracker = keras.metrics.Mean(name="reg_loss")
self.decoder = decoder
if self.decoder!=None:
self.reconstruction_loss_tracker = keras.metrics.Mean(name="reconstruction_loss")
@property
def metrics(self):
if self.decoder!=None:
return [
self.total_loss_tracker,
self.kl_loss_tracker,
self.reg_loss_tracker,
self.reconstruction_loss_tracker
]
else:
return [
self.total_loss_tracker,
self.kl_loss_tracker,
self.reg_loss_tracker,
]
def train_step(self, data):
x, target_x = data
with tf.GradientTape() as tape:
# kl loss
mu, sigma, z = self.encoder(x)
kl_loss = -0.5 * (1 + sigma - tf.square(mu) - tf.exp(sigma))
kl_loss = tf.reduce_mean(tf.reduce_sum(kl_loss, axis=1))
# Regressor
reg_prediction = self.regressor(z)
reg_loss = tf.reduce_mean(
keras.losses.mse(target_x, reg_prediction)
)
# Reconstruction
if self.decoder!=None:
reconstruction = self.decoder(z)
reconstruction_loss = tf.reduce_mean(
keras.losses.mse(x, reconstruction)
)
total_loss = kl_loss + reg_loss + reconstruction_loss
self.reconstruction_loss_tracker.update_state(reconstruction_loss)
else:
total_loss = kl_loss + reg_loss
grads = tape.gradient(total_loss, self.trainable_weights)
self.optimizer.apply_gradients(zip(grads, self.trainable_weights))
self.total_loss_tracker.update_state(total_loss)
self.kl_loss_tracker.update_state(kl_loss)
self.reg_loss_tracker.update_state(reg_loss)
return {
"loss": self.total_loss_tracker.result(),
"kl_loss": self.kl_loss_tracker.result(),
"reg_loss": self.reg_loss_tracker.result(),
}
def test_step(self, data):
x, target_x = data
# kl loss
mu, sigma, z = self.encoder(x)
kl_loss = -0.5 * (1 + sigma - tf.square(mu) - tf.exp(sigma))
kl_loss = tf.reduce_mean(tf.reduce_sum(kl_loss, axis=1))
# Regressor
reg_prediction = self.regressor(z)
reg_loss = tf.reduce_mean(
keras.losses.mse(target_x, reg_prediction)
)
# Reconstruction
if self.decoder!=None:
reconstruction = self.decoder(z)
reconstruction_loss = tf.reduce_mean(
keras.losses.mse(x, reconstruction)
)
total_loss = kl_loss + reg_loss + reconstruction_loss
else:
total_loss = kl_loss + reg_loss
return {
"loss": total_loss,
"kl_loss": kl_loss,
"reg_loss": reg_loss,
}
# Set hyperparameters
sequence_length = 200
alpha = 0.2
# Load and preprocess data
df = df # Load dataset
sensors = ['s_max_voltage_discharge', 's_min_voltage_charge', "Time_at_4.15V_s"] # Define the sensors
# Call get_data_with_kfold to get the necessary data
x_train, y_train, x_val, y_val, x_test, y_test = get_data(df1, sensors, sequence_length, alpha)
# from scipy.signal import savgol_filter
# # Apply Savitzky-Golay filter
# x_val_smoothed = savgol_filter(x_val, window_length=4, polyorder=2, axis=0)
# Setup the network parameters:
timesteps = x_train.shape[1]
input_dim = x_train.shape[2]
intermediate_dim = 32
batch_size = 256
latent_dim = 2
masking_value = -99 # used to mask values in sequences with less than 250 cycles until 250 is reached
kernel_regularizer=l1_l2(l1=0.001, l2=0.001)
dropout_rate = 0.1
# --------------------------------- Encoder --------------------------------------
inputs = Input(shape=(timesteps, input_dim,), name='encoder_input')
mask = Masking(mask_value=masking_value)(inputs)
h = Bidirectional(LSTM(intermediate_dim))(mask) # LSTM encoding
mu = Dense(latent_dim, kernel_regularizer = kernel_regularizer)(h) # VAE Z layer
mu = Dropout(dropout_rate)(mu)
sigma = Dense(latent_dim, kernel_regularizer = kernel_regularizer)(h)
sigma = Dropout(dropout_rate)(sigma)
z = Sampling()([mu, sigma])
# Instantiate the encoder model:
encoder = keras.Model(inputs, [mu, sigma, z], name='encoder')
# ------------------------------- Regressor --------------------------------------
reg_latent_inputs = Input(shape=(latent_dim,), name='z_sampling_reg')
reg_intermediate = Dense(16, activation='tanh', kernel_regularizer = kernel_regularizer)(reg_latent_inputs)
reg_intermediate = BatchNormalization()(reg_intermediate)
reg_intermediate = Dropout(dropout_rate)(reg_intermediate)
reg_outputs = Dense(1, name='reg_output', kernel_regularizer = kernel_regularizer)(reg_intermediate)
reg_outputs = Dropout(dropout_rate)(reg_outputs)
# Instantiate the classifier model:
regressor = keras.Model(reg_latent_inputs, reg_outputs, name='regressor')
print("Shape of x_train:", x_train.shape)
print("Shape of y_train:", y_train.shape)
print("Shape of x_val:", x_val.shape)
print("Shape of y_val:", y_val.shape)
print("Shape of x_test:", x_test.shape)
print("Shape of y_test:", len(y_test))Shape of x_train: (8795, 200, 3)
Shape of y_train: (8795, 1)
Shape of x_val: (1758, 200, 3)
Shape of y_val: (1758, 1)
Shape of x_test: (10, 200, 3)
Shape of y_test: 10
rve = RVE(encoder, regressor)
lr_finder = LRFinder(rve)
rve.compile(optimizer=keras.optimizers.Adam(learning_rate=0.0000001))
# with learning rate growing exponentially from 0.0000001 to 1
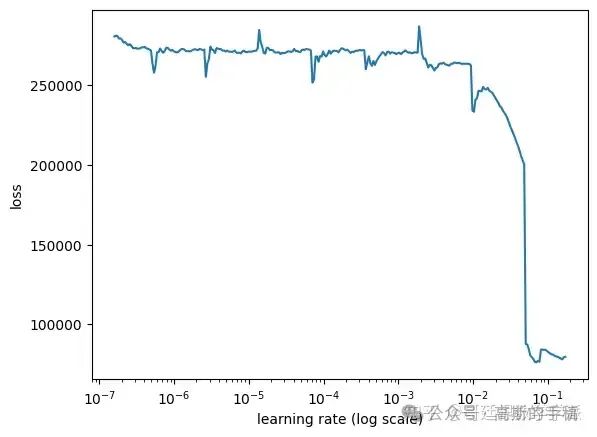
lr_finder.find(x_train, y_train, start_lr=0.0000001, end_lr=1, batch_size=batch_size, epochs=10)
# Plot the loss
lr_finder.plot_loss()
开始训练
# Instantiate the RVE model
rve = RVE(encoder, regressor)
#Compile the RVE model with the Adam optimizer
rve.compile(optimizer=keras.optimizers.Adam(0.01))
# Define the early stopping callback
early_stopping = EarlyStopping(monitor='loss', min_delta=3, patience=5, verbose=1, mode='min', restore_best_weights=True)
# Call get_data_with_kfold to get the necessary data
x_train, y_train, x_val, y_val, x_test, y_test = get_data(df1, sensors, sequence_length, alpha)
# Fit the RVE model with the callbacks
rve.fit(x_train, y_train, epochs=500, batch_size=batch_size, validation_data=(x_val, y_val), callbacks=[early_stopping])RUL估计
train_mu = viz_latent_space(rve.encoder, np.concatenate((x_train, x_val)), np.concatenate((y_train, y_val)))
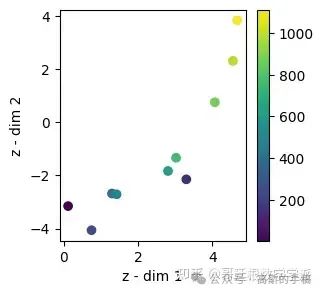
test_mu = viz_latent_space(rve.encoder, x_test, y_test)
# Evaluate
y_hat_train = rve.regressor.predict(train_mu)
y_hat_test = rve.regressor.predict(test_mu)
evaluate(np.concatenate((y_train, y_val)), y_hat_train, 'train')
evaluate(y_test, y_hat_test, 'test')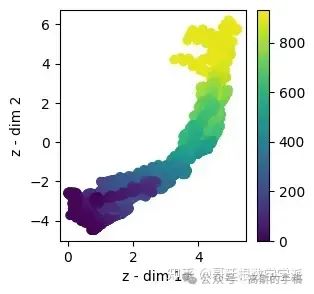
330/330 [==============================] - 0s 1ms/step 1/1 [==============================] - 0s 20ms/step train set RMSE:26.346721649169922, R2:0.9899616368349312 test set RMSE:246.51723899515346, R2:0.5192274671464132
(246.51723899515346, 0.5192274671464132)
工学博士,担任《Mechanical System and Signal Processing》《中国电机工程学报》《控制与决策》等期刊审稿专家,擅长领域:现代信号处理,机器学习,深度学习,数字孪生,时间序列分析,设备缺陷检测、设备异常检测、设备智能故障诊断与健康管理PHM等。